Cell cycle-regulated attachment of the ubiquitin-related protein SUMO to the yeast septins
- PMID: 10579719
- PMCID: PMC2169351
- DOI: 10.1083/jcb.147.5.981
Cell cycle-regulated attachment of the ubiquitin-related protein SUMO to the yeast septins
Abstract
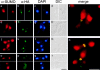
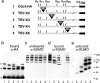
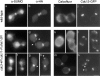
SUMO is a ubiquitin-related protein that functions as a posttranslational modification on other proteins. SUMO conjugation is essential for viability in Saccharomyces cerevisiae and is required for entry into mitosis. We have found that SUMO is attached to the septins Cdc3, Cdc11, and Shs1/Sep7 specifically during mitosis, with conjugates appearing shortly before anaphase onset and disappearing abruptly at cytokinesis. Septins are components of a belt of 10-nm filaments encircling the yeast bud neck. Intriguingly, only septins on the mother cell side of the bud neck are sumoylated. We have identified four major SUMO attachment-site lysine residues in Cdc3, one in Cdc11, and two in Shs1, all within the consensus sequence (IVL)KX(ED). Mutating these sites eliminated the vast majority of bud neck-associated SUMO, as well as the bulk of total SUMO conjugates in G(2)/M-arrested cells, indicating that sumoylated septins are the most abundant SUMO conjugates at this point in the cell cycle. This mutant has a striking defect in disassembly of septin rings, resulting in accumulation of septin rings marking previous division sites. Thus, SUMO conjugation plays a role in regulating septin ring dynamics during the cell cycle.
Figures


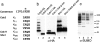


Similar articles
-
Bni5p, a septin-interacting protein, is required for normal septin function and cytokinesis in Saccharomyces cerevisiae.Mol Cell Biol. 2002 Oct;22(19):6906-20. doi: 10.1128/MCB.22.19.6906-6920.2002. Mol Cell Biol. 2002. PMID: 12215547 Free PMC article.
-
An E3-like factor that promotes SUMO conjugation to the yeast septins.Cell. 2001 Sep 21;106(6):735-44. doi: 10.1016/s0092-8674(01)00491-3. Cell. 2001. PMID: 11572779
-
Smt3, a SUMO-1 homolog, is conjugated to Cdc3, a component of septin rings at the mother-bud neck in budding yeast.Biochem Biophys Res Commun. 1999 Jun 16;259(3):582-7. doi: 10.1006/bbrc.1999.0821. Biochem Biophys Res Commun. 1999. PMID: 10364461
-
Cytoplasmic sumoylation by PIAS-type Siz1-SUMO ligase.Cell Cycle. 2008 Jun 15;7(12):1738-44. doi: 10.4161/cc.7.12.6156. Epub 2008 Jun 16. Cell Cycle. 2008. PMID: 18583943 Review.
-
Regulation of septin organization and function in yeast.Trends Cell Biol. 2003 Aug;13(8):403-9. doi: 10.1016/s0962-8924(03)00151-x. Trends Cell Biol. 2003. PMID: 12888292 Review.
Cited by
-
SUMO-1 represses apoptosis signal-regulating kinase 1 activation through physical interaction and not through covalent modification.EMBO Rep. 2005 Oct;6(10):949-55. doi: 10.1038/sj.embor.7400511. EMBO Rep. 2005. PMID: 16142216 Free PMC article.
-
An improved SUMmOn-based methodology for the identification of ubiquitin and ubiquitin-like protein conjugation sites identifies novel ubiquitin-like protein chain linkages.Proteomics. 2010 Jan;10(2):254-65. doi: 10.1002/pmic.200900648. Proteomics. 2010. PMID: 20029837 Free PMC article.
-
Septin collar formation in budding yeast requires GTP binding and direct phosphorylation by the PAK, Cla4.J Cell Biol. 2004 Mar 1;164(5):701-15. doi: 10.1083/jcb.200312070. J Cell Biol. 2004. PMID: 14993234 Free PMC article.
-
SUMO regulates the assembly and function of a cytoplasmic intermediate filament protein in C. elegans.Dev Cell. 2009 Nov;17(5):724-35. doi: 10.1016/j.devcel.2009.10.005. Dev Cell. 2009. PMID: 19922876 Free PMC article.
-
Structures of the SUMO E1 provide mechanistic insights into SUMO activation and E2 recruitment to E1.EMBO J. 2005 Feb 9;24(3):439-51. doi: 10.1038/sj.emboj.7600552. Epub 2005 Jan 20. EMBO J. 2005. PMID: 15660128 Free PMC article.
References
-
- Ausubel F.M., Brent R., Kingston R.E., Moore D.D., Smith J.A., Seidman J.G., Struhl K. Current Protocols in Molecular Biology. Wiley-Interscience; New York: 1994.
-
- Boeke J.D., LaCroute F., Fink G.R. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast5-fluoro-orotic acid resistance. Mol. Gen. Genet. 1984;197:345–346. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

