Sequential mechanism of solubilization and refolding of stable protein aggregates by a bichaperone network
- PMID: 10570141
- PMCID: PMC24133
- DOI: 10.1073/pnas.96.24.13732
Sequential mechanism of solubilization and refolding of stable protein aggregates by a bichaperone network
Abstract
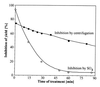
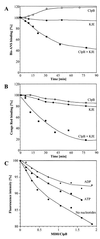
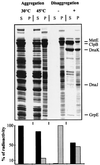
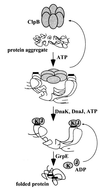
A major activity of molecular chaperones is to prevent aggregation and refold misfolded proteins. However, when allowed to form, protein aggregates are refolded poorly by most chaperones. We show here that the sequential action of two Escherichia coli chaperone systems, ClpB and DnaK-DnaJ-GrpE, can efficiently solubilize excess amounts of protein aggregates and refold them into active proteins. Measurements of aggregate turbidity, Congo red, and 4,4'-dianilino-1, 1'-binaphthyl-5,5'-disulfonic acid binding, and of the disaggregation/refolding kinetics by using a specific ClpB inhibitor, suggest a mechanism where (i) ClpB directly binds protein aggregates, ATP induces structural changes in ClpB, which (ii) increase hydrophobic exposure of the aggregates and (iii) allow DnaK-DnaJ-GrpE to bind and mediate dissociation and refolding of solubilized polypeptides into native proteins. This efficient mechanism, whereby chaperones can catalytically solubilize and refold a wide variety of large and stable protein aggregates, is a major addition to the molecular arsenal of the cell to cope with protein damage induced by stress or pathological states.
Figures

Similar articles
-
ClpB cooperates with DnaK, DnaJ, and GrpE in suppressing protein aggregation. A novel multi-chaperone system from Escherichia coli.J Biol Chem. 1999 Oct 1;274(40):28083-6. doi: 10.1074/jbc.274.40.28083. J Biol Chem. 1999. PMID: 10497158
-
Dicarboxylic amino acids and glycine-betaine regulate chaperone-mediated protein-disaggregation under stress.Mol Microbiol. 2003 Jul;49(2):401-10. doi: 10.1046/j.1365-2958.2003.03553.x. Mol Microbiol. 2003. PMID: 12828638
-
Size-dependent disaggregation of stable protein aggregates by the DnaK chaperone machinery.J Biol Chem. 2000 Jul 14;275(28):21107-13. doi: 10.1074/jbc.M001293200. J Biol Chem. 2000. PMID: 10801805
-
Interferon-gamma is a target for binding and folding by both Escherichia coli chaperone model systems GroEL/GroES and DnaK/DnaJ/GrpE.Biochimie. 1998 Aug-Sep;80(8-9):729-37. doi: 10.1016/s0300-9084(99)80026-1. Biochimie. 1998. PMID: 9865495 Review.
-
Novel insights into the mechanism of chaperone-assisted protein disaggregation.Biol Chem. 2005 Aug;386(8):739-44. doi: 10.1515/BC.2005.086. Biol Chem. 2005. PMID: 16201868 Review.
Cited by
-
Hsp110 is a bona fide chaperone using ATP to unfold stable misfolded polypeptides and reciprocally collaborate with Hsp70 to solubilize protein aggregates.J Biol Chem. 2013 Jul 19;288(29):21399-21411. doi: 10.1074/jbc.M113.479253. Epub 2013 Jun 4. J Biol Chem. 2013. PMID: 23737532 Free PMC article.
-
Evaluating EcxR for Its Possible Role in Ehrlichia chaffeensis Gene Regulation.Int J Mol Sci. 2022 Oct 22;23(21):12719. doi: 10.3390/ijms232112719. Int J Mol Sci. 2022. PMID: 36361509 Free PMC article.
-
Analysis of the cooperative ATPase cycle of the AAA+ chaperone ClpB from Thermus thermophilus by using ordered heterohexamers with an alternating subunit arrangement.J Biol Chem. 2015 Apr 10;290(15):9789-800. doi: 10.1074/jbc.M114.617696. Epub 2015 Feb 24. J Biol Chem. 2015. PMID: 25713084 Free PMC article.
-
Molecular chaperones and the assembly of the prion Sup35p, an in vitro study.EMBO J. 2006 Feb 22;25(4):822-33. doi: 10.1038/sj.emboj.7600985. Epub 2006 Feb 9. EMBO J. 2006. PMID: 16467849 Free PMC article.
-
Sequence determinants of protein aggregation: tools to increase protein solubility.Microb Cell Fact. 2005 Apr 22;4(1):11. doi: 10.1186/1475-2859-4-11. Microb Cell Fact. 2005. PMID: 15847694 Free PMC article.
References
-
- Bukau B, Horwich A L. Cell. 1998;92:351–366. - PubMed
-
- Jaenicke R. Biol Chem. 1998;379:237–243. - PubMed
-
- Turnell W G, Finch J T. J Mol Biol. 1992;227:1205–1223. - PubMed
-
- Horwich A L, Weissman J S. Cell. 1997;89:495–510. - PubMed
-
- Woo K M, Kim K I, Goldberg A L, Ha D B, Chung C H. J Biol Chem. 1992;267:20429–20434. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

