Multiple pathways for repair of DNA double-strand breaks in mammalian chromosomes
- PMID: 10567560
- PMCID: PMC84924
- DOI: 10.1128/MCB.19.12.8353
Multiple pathways for repair of DNA double-strand breaks in mammalian chromosomes
Abstract
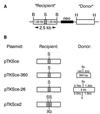
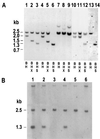
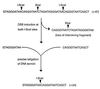
To study repair of DNA double-strand breaks (DSBs) in mammalian chromosomes, we designed DNA substrates containing a thymidine kinase (TK) gene disrupted by the 18-bp recognition site for yeast endonuclease I-SceI. Some substrates also contained a second defective TK gene sequence to serve as a genetic donor in recombinational repair. A genomic DSB was induced by introducing endonuclease I-SceI into cells containing a stably integrated DNA substrate. DSB repair was monitored by selection for TK-positive segregants. We observed that intrachromosomal DSB repair is accomplished with nearly equal efficiencies in either the presence or absence of a homologous donor sequence. DSB repair is achieved by nonhomologous end-joining or homologous recombination, but rarely by nonconservative single-strand annealing. Repair of a chromosomal DSB by homologous recombination occurs mainly by gene conversion and appears to require a donor sequence greater than a few hundred base pairs in length. Nonhomologous end-joining events typically involve loss of very few nucleotides, and some events are associated with gene amplification at the repaired locus. Additional studies revealed that precise religation of DNA ends with no other concomitant sequence alteration is a viable mode for repair of DSBs in a mammalian genome.
Figures


Similar articles
-
Induction of recombination between diverged sequences in a mammalian genome by a double-strand break.Cell Mol Life Sci. 2014 Jun;71(12):2359-71. doi: 10.1007/s00018-013-1520-0. Epub 2013 Nov 21. Cell Mol Life Sci. 2014. PMID: 24257896 Free PMC article.
-
Deletion, rearrangement, and gene conversion; genetic consequences of chromosomal double-strand breaks in human cells.Environ Mol Mutagen. 2003;42(4):288-98. doi: 10.1002/em.10201. Environ Mol Mutagen. 2003. PMID: 14673874
-
Repair of a specific double-strand break generated within a mammalian chromosome by yeast endonuclease I-SceI.Nucleic Acids Res. 1994 Dec 25;22(25):5649-57. doi: 10.1093/nar/22.25.5649. Nucleic Acids Res. 1994. PMID: 7838718 Free PMC article.
-
DSB (Im)mobility and DNA repair compartmentalization in mammalian cells.J Mol Biol. 2015 Feb 13;427(3):652-8. doi: 10.1016/j.jmb.2014.11.014. Epub 2014 Nov 24. J Mol Biol. 2015. PMID: 25463437 Review.
-
Analysis of DNA double-strand break repair pathways in mice.Mutat Res. 2007 Jan 3;614(1-2):95-108. doi: 10.1016/j.mrfmmm.2006.01.022. Epub 2006 Jun 23. Mutat Res. 2007. PMID: 16797606 Review.
Cited by
-
Manipulating the mammalian genome by homologous recombination.Proc Natl Acad Sci U S A. 2001 Jul 17;98(15):8403-10. doi: 10.1073/pnas.111009698. Proc Natl Acad Sci U S A. 2001. PMID: 11459982 Free PMC article. Review.
-
Interchromosomal gene conversion at an endogenous human cell locus.Genetics. 2001 Jun;158(2):757-67. doi: 10.1093/genetics/158.2.757. Genetics. 2001. PMID: 11404339 Free PMC article.
-
Distinct characteristics of the DNA damage response in mammalian oocytes.Exp Mol Med. 2024 Feb;56(2):319-328. doi: 10.1038/s12276-024-01178-2. Epub 2024 Feb 14. Exp Mol Med. 2024. PMID: 38355825 Free PMC article. Review.
-
Both the charged linker region and ATPase domain of Hsp90 are essential for Rad51-dependent DNA repair.Eukaryot Cell. 2015 Jan;14(1):64-77. doi: 10.1128/EC.00159-14. Epub 2014 Nov 7. Eukaryot Cell. 2015. PMID: 25380755 Free PMC article.
-
Variants of the human RAD52 gene confer defects in ionizing radiation resistance and homologous recombination repair in budding yeast.Microb Cell. 2020 Jul 20;7(10):270-285. doi: 10.15698/mic2020.10.732. Microb Cell. 2020. PMID: 33015141 Free PMC article.
References
-
- Bernstein C, Bernstein H. Aging, sex, and DNA repair. San Diego, Calif: Academic Press, Inc.; 1991.
-
- Bollag R J, Waldman A S, Liskay R M. Homologous recombination in mammalian cells. Annu Rev Genet. 1989;23:199–225. - PubMed
-
- Camerini-Otero R D, Hsieh P. Homologous recombination proteins in prokaryotes and eukaryotes. Annu Rev Genet. 1995;29:509–552. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
