A ligand binding domain mutation in the mouse glucocorticoid receptor functionally links chromatin remodeling and transcription initiation
- PMID: 10567540
- PMCID: PMC84899
- DOI: 10.1128/MCB.19.12.8146
A ligand binding domain mutation in the mouse glucocorticoid receptor functionally links chromatin remodeling and transcription initiation
Abstract
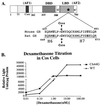
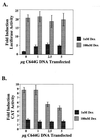
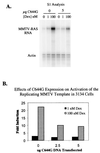
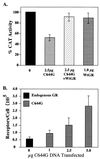
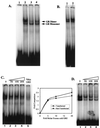
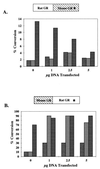
We utilized the mouse mammary tumor virus (MMTV) long terminal repeat (LTR) in vivo to understand how the interaction of the glucocorticoid receptor (GR) with a nucleosome-assembled promoter allows access of factors required for the transition from a repressed promoter to a derepressed, transcriptionally competent promoter. A mutation (C644G) in the ligand binding domain (LBD) of the mouse GR has provided information regarding the steps required in the derepression/activation process and in the functional significance of the two major transcriptional activation domains, AF1 and AF2. The mutant GR activates transcription from a transiently transfected promoter that has a disordered nucleosomal structure, though significantly less well than the wild-type GR. With an integrated, replicated promoter, which is assembled in an ordered nucleosomal array, the mutant GR does not activate transcription, and it fails to induce chromatin remodeling of the MMTV LTR promoter, as indicated by nuclease accessibility assays. Together, these findings support a two-step model for the transition of a nucleosome-assembled, repressed promoter to its transcriptionally active, derepressed form. In addition, we find that the C-terminal GR mutation is dominant over the transcription activation function of the N-terminal GR activation domain. These findings suggest that the primary activation function of the C-terminal activation domain is different from the function of the N-terminal activation domain and that it is required for derepression of the chromatin-repressed MMTV promoter.
Figures

Similar articles
-
Glucocorticoid receptor domain requirements for chromatin remodeling and transcriptional activation of the mouse mammary tumor virus promoter in different nucleoprotein contexts.J Biol Chem. 2002 Aug 2;277(31):28247-55. doi: 10.1074/jbc.M203898200. Epub 2002 May 23. J Biol Chem. 2002. PMID: 12029095
-
Identification of glucocorticoid receptor domains necessary for transcriptional activation of the mouse mammary tumor virus promoter integrated in the genome.Exp Cell Res. 1998 Mar 15;239(2):454-62. doi: 10.1006/excr.1997.3920. Exp Cell Res. 1998. PMID: 9521864
-
Dynamics of gene targeting and chromatin remodelling by nuclear receptors.Biochem Soc Trans. 2000;28(4):405-10. Biochem Soc Trans. 2000. PMID: 10961929
-
Glucocorticoid receptor-mediated chromatin remodeling in vivo.Oncogene. 2001 May 28;20(24):3039-46. doi: 10.1038/sj.onc.1204328. Oncogene. 2001. PMID: 11420719 Review.
-
Chromatin remodeling during glucocorticoid receptor regulated transactivation.Biochim Biophys Acta. 2012 Jul;1819(7):716-26. doi: 10.1016/j.bbagrm.2012.02.019. Epub 2012 Mar 6. Biochim Biophys Acta. 2012. PMID: 22425674 Free PMC article. Review.
Cited by
-
The Mediator subunit MED1/TRAP220 is required for optimal glucocorticoid receptor-mediated transcription activation.Nucleic Acids Res. 2007;35(18):6161-9. doi: 10.1093/nar/gkm661. Epub 2007 Sep 7. Nucleic Acids Res. 2007. PMID: 17827210 Free PMC article.
-
Crucial roles for interactions between MLL3/4 and INI1 in nuclear receptor transactivation.Mol Endocrinol. 2009 May;23(5):610-9. doi: 10.1210/me.2008-0455. Epub 2009 Feb 12. Mol Endocrinol. 2009. PMID: 19221051 Free PMC article.
-
Trichostatin A inhibits beta-casein expression in mammary epithelial cells.J Cell Biochem. 2001;83(4):660-70. doi: 10.1002/jcb.1260. J Cell Biochem. 2001. PMID: 11746508 Free PMC article.
-
Transcriptional effects of glucocorticoid receptors in the dentate gyrus increase anxiety-related behaviors.PLoS One. 2009 Nov 2;4(11):e7704. doi: 10.1371/journal.pone.0007704. PLoS One. 2009. PMID: 19888328 Free PMC article.
-
Expression level-dependent contribution of glucocorticoid receptor domains for functional interaction with STAT5.Mol Cell Biol. 2001 May;21(9):3266-79. doi: 10.1128/MCB.21.9.3266-3279.2001. Mol Cell Biol. 2001. PMID: 11287629 Free PMC article.
References
-
- Almhöf T, Wallberg A E, Gustafsson J-Å, Wright A P H. Role of important hydrophobic amino acids in the interaction between the glucocorticoid receptor t1-core activation domain and target factors. Biochemistry. 1998;37:9586–9594. - PubMed
-
- Archer T K, Cordingley M G, Marsaud V, Richard-Foy H, Hager G L. Proceedings: Second International CBT Symposium on the Steroid/Thyroid Receptor Family and Gene Regulation. Berlin: Birkhauser Verlag AG; 1989. pp. 221–238.
-
- Archer T K, Lee H-L, Cordingley M G, Mymryk J S, Fragoso G, Berard D S, Hager G L. Differential steroid hormone induction of transcription from the mouse mammary tumor virus promoter. Mol Endocrinol. 1994;8:568–576. - PubMed
-
- Archer T K, Lefebvre P, Wolford R G, Hager G L. Transcription factor loading on the MMTV promoter: a bimodal mechanism for promoter activation. Science. 1992;255:1573–1576. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
