Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress
- PMID: 10564271
- PMCID: PMC25679
- DOI: 10.1091/mbc.10.11.3787
Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress
Abstract
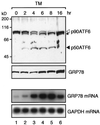
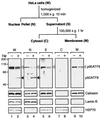
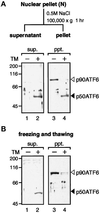
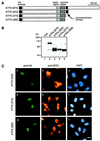
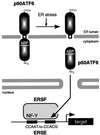
The unfolded protein response (UPR) controls the levels of molecular chaperones and enzymes involved in protein folding in the endoplasmic reticulum (ER). We recently isolated ATF6 as a candidate for mammalian UPR-specific transcription factor. We report here that ATF6 constitutively expressed as a 90-kDa protein (p90ATF6) is directly converted to a 50-kDa protein (p50ATF6) in ER-stressed cells. Furthermore, we showed that the most important consequence of this conversion was altered subcellular localization; p90ATF6 is embedded in the ER, whereas p50ATF6 is a nuclear protein. p90ATF6 is a type II transmembrane glycoprotein with a hydrophobic stretch in the middle of the molecule. Thus, the N-terminal half containing a basic leucine zipper motif is oriented facing the cytoplasm. Full-length ATF6 as well as its C-terminal deletion mutant carrying the transmembrane domain is localized in the ER when transfected. In contrast, mutant ATF6 representing the cytoplasmic region translocates into the nucleus and activates transcription of the endogenous GRP78/BiP gene. We propose that ER stress-induced proteolysis of membrane-bound p90ATF6 releases soluble p50ATF6, leading to induced transcription in the nucleus. Unlike yeast UPR, mammalian UPR appears to use a system similar to that reported for cholesterol homeostasis.
Figures




Similar articles
-
Identification of the G13 (cAMP-response-element-binding protein-related protein) gene product related to activating transcription factor 6 as a transcriptional activator of the mammalian unfolded protein response.Biochem J. 2001 Apr 1;355(Pt 1):19-28. doi: 10.1042/0264-6021:3550019. Biochem J. 2001. PMID: 11256944 Free PMC article.
-
ATF6 activated by proteolysis binds in the presence of NF-Y (CBF) directly to the cis-acting element responsible for the mammalian unfolded protein response.Mol Cell Biol. 2000 Sep;20(18):6755-67. doi: 10.1128/MCB.20.18.6755-6767.2000. Mol Cell Biol. 2000. PMID: 10958673 Free PMC article.
-
Underglycosylation of ATF6 as a novel sensing mechanism for activation of the unfolded protein response.J Biol Chem. 2004 Mar 19;279(12):11354-63. doi: 10.1074/jbc.M309804200. Epub 2003 Dec 29. J Biol Chem. 2004. PMID: 14699159
-
[The regulation of unfolded protein response by OASIS, a transmembrane bZIP transcription factor, in astrocytes].Nihon Yakurigaku Zasshi. 2004 Dec;124(6):383-90. doi: 10.1254/fpj.124.383. Nihon Yakurigaku Zasshi. 2004. PMID: 15572842 Review. Japanese.
-
Physiological unfolded protein response regulated by OASIS family members, transmembrane bZIP transcription factors.IUBMB Life. 2011 Apr;63(4):233-9. doi: 10.1002/iub.433. Epub 2011 Mar 24. IUBMB Life. 2011. PMID: 21438114 Review.
Cited by
-
Bacteria, the endoplasmic reticulum and the unfolded protein response: friends or foes?Nat Rev Microbiol. 2015 Feb;13(2):71-82. doi: 10.1038/nrmicro3393. Epub 2014 Dec 15. Nat Rev Microbiol. 2015. PMID: 25534809 Free PMC article. Review.
-
Relevance of Endoplasmic Reticulum Stress Cell Signaling in Liver Cold Ischemia Reperfusion Injury.Int J Mol Sci. 2016 May 25;17(6):807. doi: 10.3390/ijms17060807. Int J Mol Sci. 2016. PMID: 27231901 Free PMC article. Review.
-
Limitation of individual folding resources in the ER leads to outcomes distinct from the unfolded protein response.J Cell Sci. 2012 Oct 15;125(Pt 20):4865-75. doi: 10.1242/jcs.108928. Epub 2012 Aug 1. J Cell Sci. 2012. PMID: 22854046 Free PMC article.
-
Mutations in the unfolded protein response regulator ATF6 cause the cone dysfunction disorder achromatopsia.Nat Genet. 2015 Jul;47(7):757-65. doi: 10.1038/ng.3319. Epub 2015 Jun 1. Nat Genet. 2015. PMID: 26029869 Free PMC article.
-
A mathematical model of the unfolded protein stress response reveals the decision mechanism for recovery, adaptation and apoptosis.BMC Syst Biol. 2013 Feb 21;7:16. doi: 10.1186/1752-0509-7-16. BMC Syst Biol. 2013. PMID: 23433609 Free PMC article.
References
-
- Brown MS, Goldstein JL. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331–340. - PubMed
-
- Chan Y-M, Jan YN. Roles for proteolysis and trafficking in Notch maturation and signal transduction. Cell. 1998;94:423–426. - PubMed
-
- Chapman RE, Walter P. Translational attenuation mediated by an mRNA intron. Curr Biol. 1997;7:850–859. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

