A proinflammatory cytokine inhibits p53 tumor suppressor activity
- PMID: 10562313
- PMCID: PMC2195698
- DOI: 10.1084/jem.190.10.1375
A proinflammatory cytokine inhibits p53 tumor suppressor activity
Abstract
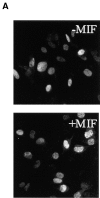
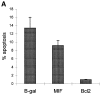
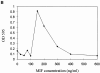
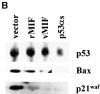
p53 has a key role in the negative regulation of cell proliferation, in the maintenance of genomic stability, and in the suppression of transformation and tumorigenesis. To identify novel regulators of p53, we undertook two functional screens to isolate genes which bypassed either p53-mediated growth arrest or apoptosis. In both screens, we isolated cDNAs encoding macrophage migration inhibitory factor (MIF), a cytokine that was shown previously to exert both local and systemic proinflammatory activities. Treatment with MIF overcame p53 activity in three different biological assays, and suppressed its activity as a transcriptional activator. The observation that a proinflammatory cytokine, MIF, is capable of functionally inactivating a tumor suppressor, p53, may provide a link between inflammation and tumorigenesis.
Figures











Comment in
-
At the crossroads of inflammation and tumorigenesis.J Exp Med. 1999 Nov 15;190(10):1367-70. doi: 10.1084/jem.190.10.1367. J Exp Med. 1999. PMID: 10562311 Free PMC article. No abstract available.
Similar articles
-
At the crossroads of inflammation and tumorigenesis.J Exp Med. 1999 Nov 15;190(10):1367-70. doi: 10.1084/jem.190.10.1367. J Exp Med. 1999. PMID: 10562311 Free PMC article. No abstract available.
-
The p53-dependent effects of macrophage migration inhibitory factor revealed by gene targeting.Proc Natl Acad Sci U S A. 2003 Aug 5;100(16):9354-9. doi: 10.1073/pnas.1533295100. Epub 2003 Jul 23. Proc Natl Acad Sci U S A. 2003. PMID: 12878730 Free PMC article.
-
Macrophage migration inhibitory factor (MIF) sustains macrophage proinflammatory function by inhibiting p53: regulatory role in the innate immune response.Proc Natl Acad Sci U S A. 2002 Jan 8;99(1):345-50. doi: 10.1073/pnas.012511599. Epub 2001 Dec 26. Proc Natl Acad Sci U S A. 2002. PMID: 11756671 Free PMC article.
-
Control of p53 and NF-κB signaling by WIP1 and MIF: role in cellular senescence and organismal aging.Cell Signal. 2011 May;23(5):747-52. doi: 10.1016/j.cellsig.2010.10.012. Epub 2010 Oct 16. Cell Signal. 2011. PMID: 20940041 Review.
-
Inflammation and cancer: macrophage migration inhibitory factor (MIF)--the potential missing link.QJM. 2010 Nov;103(11):831-6. doi: 10.1093/qjmed/hcq148. Epub 2010 Aug 30. QJM. 2010. PMID: 20805118 Free PMC article. Review.
Cited by
-
Macrophage migration inhibitory factor produced by the tumour stroma but not by tumour cells regulates angiogenesis in the B16-F10 melanoma model.Br J Cancer. 2012 Oct 23;107(9):1498-505. doi: 10.1038/bjc.2012.392. Epub 2012 Sep 6. Br J Cancer. 2012. PMID: 22955855 Free PMC article.
-
Discovering the route from inflammation to pancreatic cancer.Minerva Gastroenterol Dietol. 2012 Dec;58(4):283-97. Minerva Gastroenterol Dietol. 2012. PMID: 23207606 Free PMC article. Review.
-
Plasmodium yoelii macrophage migration inhibitory factor is necessary for efficient liver-stage development.Infect Immun. 2012 Apr;80(4):1399-407. doi: 10.1128/IAI.05861-11. Epub 2012 Jan 17. Infect Immun. 2012. PMID: 22252874 Free PMC article.
-
Role of MIF Cytokine/CD74 Receptor Pathway in Protecting Against Injury and Promoting Repair.Front Immunol. 2020 Jun 23;11:1273. doi: 10.3389/fimmu.2020.01273. eCollection 2020. Front Immunol. 2020. PMID: 32655566 Free PMC article. Review.
-
OxMIF: a druggable isoform of macrophage migration inhibitory factor in cancer and inflammatory diseases.J Immunother Cancer. 2022 Sep;10(9):e005475. doi: 10.1136/jitc-2022-005475. J Immunother Cancer. 2022. PMID: 36180072 Free PMC article. Review.
References
-
- Ko L.J., Prives C. p53puzzle and paradigm. Genes Dev. 1996;10:1054–1072. - PubMed
-
- Hansen R., Oren M. p53from inductive signal to cellular effect. Curr. Opin. Genet. Dev. 1997;7:46–51. - PubMed
-
- Kastan M.B., Onyekwere O., Sidransky D., Vogelstein B., Craig R.W. Participation of p53 protein in the cellular response to DNA damage. Cancer Res. 1991;51:6304–6311. - PubMed
-
- Yonish-Rouach E., Resnitzky D., Lotem J., Sachs L., Kimchi A., Oren M. Wild-type p53 induces apoptosis of myeloid leukaemic cells that is inhibited by interleukin-6. Nature. 1991;353:345–347. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

