Division versus fusion: Dnm1p and Fzo1p antagonistically regulate mitochondrial shape
- PMID: 10562274
- PMCID: PMC2156171
- DOI: 10.1083/jcb.147.4.699
Division versus fusion: Dnm1p and Fzo1p antagonistically regulate mitochondrial shape
Abstract
In yeast, mitochondrial division and fusion are highly regulated during growth, mating and sporulation, yet the mechanisms controlling these activities are unknown. Using a novel screen, we isolated mutants in which mitochondria lose their normal structure, and instead form a large network of interconnected tubules. These mutants, which appear defective in mitochondrial division, all carried mutations in DNM1, a dynamin-related protein that localizes to mitochondria. We also isolated mutants containing numerous mitochondrial fragments. These mutants were defective in FZO1, a gene previously shown to be required for mitochondrial fusion. Surprisingly, we found that in dnm1 fzo1 double mutants, normal mitochondrial shape is restored. Induction of Dnm1p expression in dnm1 fzo1 cells caused rapid fragmentation of mitochondria. We propose that dnm1 mutants are defective in the mitochondrial division, an activity antagonistic to fusion. Our results thus suggest that mitochondrial shape is normally controlled by a balance between division and fusion which requires Dnm1p and Fzo1p, respectively.
Figures
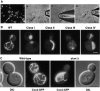
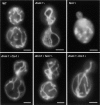
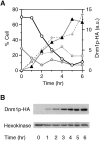
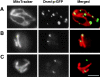

Similar articles
-
The dynamin-related GTPase Dnm1 regulates mitochondrial fission in yeast.Nat Cell Biol. 1999 Sep;1(5):298-304. doi: 10.1038/13014. Nat Cell Biol. 1999. PMID: 10559943 Free PMC article.
-
UGO1 encodes an outer membrane protein required for mitochondrial fusion.J Cell Biol. 2001 Mar 19;152(6):1123-34. doi: 10.1083/jcb.152.6.1123. J Cell Biol. 2001. PMID: 11257114 Free PMC article.
-
The intramitochondrial dynamin-related GTPase, Mgm1p, is a component of a protein complex that mediates mitochondrial fusion.J Cell Biol. 2003 Feb 3;160(3):303-11. doi: 10.1083/jcb.200209015. J Cell Biol. 2003. PMID: 12566426 Free PMC article.
-
Yeast mitochondrial dynamics: fusion, division, segregation, and shape.Microsc Res Tech. 2000 Dec 15;51(6):573-83. doi: 10.1002/1097-0029(20001215)51:6<573::AID-JEMT7>3.0.CO;2-2. Microsc Res Tech. 2000. PMID: 11169859 Review.
-
Mitochondrial dynamics in yeast.Annu Rev Cell Dev Biol. 1998;14:265-303. doi: 10.1146/annurev.cellbio.14.1.265. Annu Rev Cell Dev Biol. 1998. PMID: 9891785 Review.
Cited by
-
Mitochondrial dynamics in neurodegeneration.Trends Cell Biol. 2013 Feb;23(2):64-71. doi: 10.1016/j.tcb.2012.10.006. Epub 2012 Nov 16. Trends Cell Biol. 2013. PMID: 23159640 Free PMC article. Review.
-
Regulation of mitochondrial dynamics: convergences and divergences between yeast and vertebrates.Cell Mol Life Sci. 2013 Mar;70(6):951-76. doi: 10.1007/s00018-012-1066-6. Epub 2012 Jul 18. Cell Mol Life Sci. 2013. PMID: 22806564 Free PMC article. Review.
-
MTCH2 cooperates with MFN2 and lysophosphatidic acid synthesis to sustain mitochondrial fusion.EMBO Rep. 2024 Jan;25(1):45-67. doi: 10.1038/s44319-023-00009-1. Epub 2023 Dec 14. EMBO Rep. 2024. PMID: 38177900 Free PMC article.
-
Mitochondrial dynamics in macrophages: divide to conquer or unite to survive?Biochem Soc Trans. 2023 Feb 27;51(1):41-56. doi: 10.1042/BST20220014. Biochem Soc Trans. 2023. PMID: 36815717 Free PMC article.
-
A novel mitochondrial ubiquitin ligase plays a critical role in mitochondrial dynamics.EMBO J. 2006 Aug 9;25(15):3618-26. doi: 10.1038/sj.emboj.7601249. Epub 2006 Jul 27. EMBO J. 2006. PMID: 16874301 Free PMC article.
References
-
- Adams A., Gottschling D., Kaiser C., Stearns T. Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press; Plainview, NY : 1997.
-
- Bereiter-Hahn J., Voth M. Dynamics of mitochondria in living cellsshape changes, dislocations, fusion, and fission of mitochondria. Microsc. Res. Tech. 1994;27:198–219 . - PubMed
-
- Brachmann C.B., Davies A., Caputo E., Li J., Hieter P., Boeke J.D. Designer deletion strains from Saccharomyces cerevisiae 288Ca useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132 . - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

