The dynamin-related GTPase Dnm1 regulates mitochondrial fission in yeast
- PMID: 10559943
- PMCID: PMC3739991
- DOI: 10.1038/13014
The dynamin-related GTPase Dnm1 regulates mitochondrial fission in yeast
Abstract
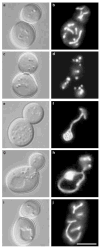
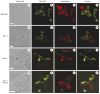
The dynamin-related GTPase Dnm1 controls mitochondrial morphology in yeast. Here we show that dnm1 mutations convert the mitochondrial compartment into a planar 'net' of interconnected tubules. We propose that this net morphology results from a defect in mitochondrial fission. Immunogold labelling localizes Dnm1 to the cytoplasmic face of constricted mitochondrial tubules that appear to be dividing and to the ends of mitochondrial tubules that appear to have recently completed division. The activity of Dnm1 is epistatic to that of Fzo1, a GTPase in the outer mitochondrial membrane that regulates mitochondrial fusion. dnm1 mutations prevent mitochondrial fragmentation in fzo1 mutant strains. These findings indicate that Dnm1 regulates mitochondrial fission, assembling on the cytoplasmic face of mitochondrial tubules at sites at which division will occur.
Figures
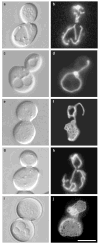
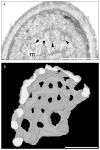
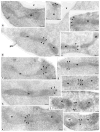

Similar articles
-
Division versus fusion: Dnm1p and Fzo1p antagonistically regulate mitochondrial shape.J Cell Biol. 1999 Nov 15;147(4):699-706. doi: 10.1083/jcb.147.4.699. J Cell Biol. 1999. PMID: 10562274 Free PMC article.
-
The intramitochondrial dynamin-related GTPase, Mgm1p, is a component of a protein complex that mediates mitochondrial fusion.J Cell Biol. 2003 Feb 3;160(3):303-11. doi: 10.1083/jcb.200209015. J Cell Biol. 2003. PMID: 12566426 Free PMC article.
-
The dynamin-related GTPase, Mgm1p, is an intermembrane space protein required for maintenance of fusion competent mitochondria.J Cell Biol. 2000 Oct 16;151(2):341-52. doi: 10.1083/jcb.151.2.341. J Cell Biol. 2000. PMID: 11038181 Free PMC article.
-
Mitochondrial dynamics in yeast.Annu Rev Cell Dev Biol. 1998;14:265-303. doi: 10.1146/annurev.cellbio.14.1.265. Annu Rev Cell Dev Biol. 1998. PMID: 9891785 Review.
-
Fusion, fragmentation, and fission of mitochondria.Biochemistry (Mosc). 2003 Aug;68(8):838-49. doi: 10.1023/a:1025738712958. Biochemistry (Mosc). 2003. PMID: 12948383 Review.
Cited by
-
Molecular mechanisms of mitochondrial dynamics.Nat Rev Mol Cell Biol. 2024 Oct 17. doi: 10.1038/s41580-024-00785-1. Online ahead of print. Nat Rev Mol Cell Biol. 2024. PMID: 39420231 Review.
-
Ribosomes hibernate on mitochondria during cellular stress.Nat Commun. 2024 Oct 8;15(1):8666. doi: 10.1038/s41467-024-52911-4. Nat Commun. 2024. PMID: 39379376 Free PMC article.
-
Dynamins combine mechano-constriction and membrane remodeling to enable two-step mitochondrial fission via a 'snap-through' instability.bioRxiv [Preprint]. 2024 Aug 20:2024.08.19.608723. doi: 10.1101/2024.08.19.608723. bioRxiv. 2024. PMID: 39229060 Free PMC article. Preprint.
-
Atg44/Mdi1/mitofissin facilitates Dnm1-mediated mitochondrial fission.Autophagy. 2024 Oct;20(10):2314-2322. doi: 10.1080/15548627.2024.2360345. Epub 2024 Jun 4. Autophagy. 2024. PMID: 38818923 Free PMC article.
-
Mitochondria-ER-PM contacts regulate mitochondrial division and PI(4)P distribution.J Cell Biol. 2024 Sep 2;223(9):e202308144. doi: 10.1083/jcb.202308144. Epub 2024 May 23. J Cell Biol. 2024. PMID: 38781029
References
-
- Williamson DH, et al. In: Genetics, Biogenesis and Bioenergetics of Mitochondria. Bandlow W, Schweyen RJ, Thomas DY, Wolf K, Kaudewitz F, editors. Walter de Gruyter; Berlin: 1976. pp. 99–115.
-
- Green DR, Reed JC. Mitochondria and apoptosis. Science. 1998;281:1309–1312. - PubMed
-
- Tourte M, Mignotte F, Mounolou JC. Organization and replicative activity of the mitochondria of oogenic and previtellogenic oocytes in Xenopus laevis. Dev Growth Differ. 1981;23:9–21. - PubMed
-
- Fuller MT. In: The Development of Drosophila melanogaster. Bate M, Martinez-Arias A, editors. Cold Spring Harb. Lab. Press; Cold Spring Harbor: 1993. pp. 71–147.
-
- Bereiter-Hahn J. Behavior of mitochondria in the living cell. Int Rev Cytol. 1990;122:1–63. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

