hMutSalpha- and hMutLalpha-dependent phosphorylation of p53 in response to DNA methylator damage
- PMID: 10535931
- PMCID: PMC22926
- DOI: 10.1073/pnas.96.22.12384
hMutSalpha- and hMutLalpha-dependent phosphorylation of p53 in response to DNA methylator damage
Abstract
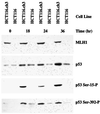
hMSH2.hMSH6 heterodimer (hMutSalpha) and hMLH1.hPMS2 complex (hMutLalpha) have been implicated in the cytotoxic response of mammalian cells to a number of DNA-damaging compounds, including methylating agents that produce O(6)-methylguanine (O(6)MeG) adducts. This study demonstrates that O(6)MeG lesions, in which the damaged base is paired with either T or C, are subject to excision repair in a reaction that depends on a functional mismatch repair system. Furthermore, treatment of human cells with the S(N)1 DNA methylators N-methyl-N-nitrosourea or N-methyl-N'-nitro-N-nitrosoguanidine results in p53 phosphorylation on serine residues 15 and 392, and these phosphorylation events depend on the presence of functional hMutSalpha and hMutLalpha. Coupled with the previous demonstration that O(6)MeG.T and O(6)MeG.C pairs are recognized by hMutSalpha, these results implicate action of the mismatch repair system in the initial step of a damage-signaling cascade that can lead to cell-cycle checkpoint activation or cell death in response to DNA methylator damage.
Figures


Similar articles
-
N-terminus of hMLH1 confers interaction of hMutLalpha and hMutLbeta with hMutSalpha.Nucleic Acids Res. 2003 Jun 15;31(12):3217-26. doi: 10.1093/nar/gkg420. Nucleic Acids Res. 2003. PMID: 12799449 Free PMC article.
-
Phosphorylated hMSH6: DNA mismatch versus DNA damage recognition.Mutat Res. 2011 Jan 10;706(1-2):36-45. doi: 10.1016/j.mrfmmm.2010.10.008. Epub 2010 Oct 28. Mutat Res. 2011. PMID: 21035467 Free PMC article.
-
hMutSalpha forms an ATP-dependent complex with hMutLalpha and hMutLbeta on DNA.Nucleic Acids Res. 2002 Feb 1;30(3):711-8. doi: 10.1093/nar/30.3.711. Nucleic Acids Res. 2002. PMID: 11809883 Free PMC article.
-
Structural, molecular and cellular functions of MSH2 and MSH6 during DNA mismatch repair, damage signaling and other noncanonical activities.Mutat Res. 2013 Mar-Apr;743-744:53-66. doi: 10.1016/j.mrfmmm.2012.12.008. Epub 2013 Feb 4. Mutat Res. 2013. PMID: 23391514 Free PMC article. Review.
-
The role of mismatch repair in DNA damage-induced apoptosis.Oncol Res. 1999;11(9):393-400. Oncol Res. 1999. PMID: 10821533 Review.
Cited by
-
High rate of CAD gene amplification in human cells deficient in MLH1 or MSH6.Proc Natl Acad Sci U S A. 2001 Nov 20;98(24):13802-7. doi: 10.1073/pnas.241508098. Proc Natl Acad Sci U S A. 2001. PMID: 11717437 Free PMC article.
-
DNA mismatch repair in eukaryotes and bacteria.J Nucleic Acids. 2010 Jul 27;2010:260512. doi: 10.4061/2010/260512. J Nucleic Acids. 2010. PMID: 20725617 Free PMC article.
-
Increased sensitivity of p53-deficient cells to anticancer agents due to loss of Pms2.Br J Cancer. 2002 Oct 21;87(9):1027-33. doi: 10.1038/sj.bjc.6600599. Br J Cancer. 2002. PMID: 12434296 Free PMC article.
-
MSH2 and ATR form a signaling module and regulate two branches of the damage response to DNA methylation.Proc Natl Acad Sci U S A. 2003 Dec 23;100(26):15387-92. doi: 10.1073/pnas.2536810100. Epub 2003 Dec 3. Proc Natl Acad Sci U S A. 2003. PMID: 14657349 Free PMC article.
-
Methylator-induced, mismatch repair-dependent G2 arrest is activated through Chk1 and Chk2.Mol Biol Cell. 2005 Mar;16(3):1513-26. doi: 10.1091/mbc.e04-02-0089. Epub 2005 Jan 12. Mol Biol Cell. 2005. PMID: 15647386 Free PMC article.
References
-
- Branch P, Aquilina G, Bignami M, Karran P. Nature (London) 1993;362:652–654. - PubMed
-
- Koi M, Umar A, Chauhan D P, Cherian S P, Carethers J M, Kunkel T A, Boland C R. Cancer Res. 1994;54:4308–4312. - PubMed
-
- de Wind N, Dekker M, Berns A, Radman M, te Riele H. Cell. 1995;82:321–330. - PubMed
-
- Drummond J T, Anthoney A, Brown R, Modrich P. J Biol Chem. 1996;271:19645–19648. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

