Reciprocal-space solvent flattening
- PMID: 10531484
- PMCID: PMC2745881
- DOI: 10.1107/s0907444999010033
Reciprocal-space solvent flattening
Abstract
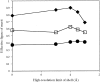
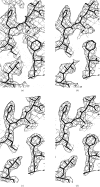
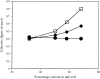
Solvent flattening is a powerful tool for improving crystallographic phases for macromolecular structures obtained at moderate resolution, but uncertainties in the optimal weighting of experimental phases and modified phases make it difficult to extract all the phase information possible. Solvent flattening is essentially an iterative method for maximizing a likelihood function which consists of (i) experimental phase information and (ii) information on the likelihood of various arrangements of electron density in a map, but the likelihood function is generally not explicitly defined. In this work, a procedure is described for reciprocal-space maximization of a likelihood function based on experimental phases and characteristics of the electron-density map. The procedure can readily be applied to phase improvement based on solvent flattening and can potentially incorporate information on a wide variety of other characteristics of the electron-density map.
Figures
Similar articles
-
Evaluation of macromolecular electron-density map quality using the correlation of local r.m.s. density.Acta Crystallogr D Biol Crystallogr. 1999 Nov;55(Pt 11):1872-7. doi: 10.1107/s090744499901029x. Acta Crystallogr D Biol Crystallogr. 1999. PMID: 10531485 Free PMC article.
-
Entropy maximization constrained by solvent flatness: a new method for macromolecular phase extension and map improvement.Acta Crystallogr D Biol Crystallogr. 1993 Jan 1;49(Pt 1):193-212. doi: 10.1107/S0907444992008540. Acta Crystallogr D Biol Crystallogr. 1993. PMID: 15299561
-
The use of wavelet transforms in low-resolution phase extension.Acta Crystallogr D Biol Crystallogr. 2000 May;56(Pt 5):625-33. doi: 10.1107/s0907444900003103. Acta Crystallogr D Biol Crystallogr. 2000. PMID: 10771432
-
Rosetta Structure Prediction as a Tool for Solving Difficult Molecular Replacement Problems.Methods Mol Biol. 2017;1607:455-466. doi: 10.1007/978-1-4939-7000-1_19. Methods Mol Biol. 2017. PMID: 28573585 Review.
-
Ab initio phase determination and phase extension using non-crystallographic symmetry.Curr Opin Struct Biol. 1995 Oct;5(5):650-5. doi: 10.1016/0959-440x(95)80058-1. Curr Opin Struct Biol. 1995. PMID: 8574701 Review.
Cited by
-
Robust and automatic beamstop shadow outlier rejection: combining crystallographic statistics with modern clustering under a semi-supervised learning strategy.Acta Crystallogr D Struct Biol. 2024 Oct 1;80(Pt 10):722-732. doi: 10.1107/S2059798324008519. Epub 2024 Oct 1. Acta Crystallogr D Struct Biol. 2024. PMID: 39361355 Free PMC article.
-
The Automatic Solution of Macromolecular Crystal Structures via Molecular Replacement Techniques: REMO22 and Its Pipeline.Int J Mol Sci. 2023 Mar 23;24(7):6070. doi: 10.3390/ijms24076070. Int J Mol Sci. 2023. PMID: 37047043 Free PMC article.
-
A microbial transporter of the dietary antioxidant ergothioneine.Cell. 2022 Nov 23;185(24):4526-4540.e18. doi: 10.1016/j.cell.2022.10.008. Epub 2022 Nov 7. Cell. 2022. PMID: 36347253 Free PMC article.
-
Structures of diverse poxin cGAMP nucleases reveal a widespread role for cGAS-STING evasion in host-pathogen conflict.Elife. 2020 Nov 16;9:e59753. doi: 10.7554/eLife.59753. Elife. 2020. PMID: 33191912 Free PMC article.
-
Improvement of cryo-EM maps by density modification.Nat Methods. 2020 Sep;17(9):923-927. doi: 10.1038/s41592-020-0914-9. Epub 2020 Aug 17. Nat Methods. 2020. PMID: 32807957 Free PMC article.
References
-
- Abrahams, J. P. & Leslie, A. G. W. (1996). Acta Cryst. D52, 30–32. - PubMed
-
- American Type Culture Collection (1992). Catalogue of Bacteria and Bacteriophages, 18th ed., pp. 271–272.
-
- Baker, D., Krukowski, A. E. & Agard, D. A. (1993). Acta Cryst. D49, 186–192. - PubMed
-
- Box, G. E. P. & Tiao, G. C. (1973). Bayesian Inference in Statistical Analysis. New York: John Wiley.
-
- Bricogne, G. (1984). Acta Cryst. A40, 410–445.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

