Formation process of autophagosome is traced with Apg8/Aut7p in yeast
- PMID: 10525546
- PMCID: PMC2174223
- DOI: 10.1083/jcb.147.2.435
Formation process of autophagosome is traced with Apg8/Aut7p in yeast
Abstract
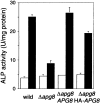
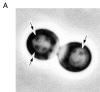
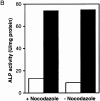
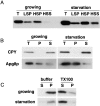
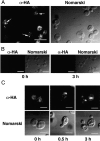
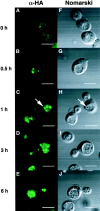
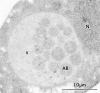
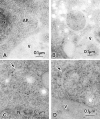
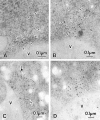
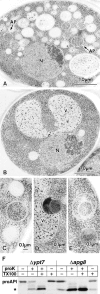
We characterized Apg8/Aut7p essential for autophagy in yeast. Apg8p was transcriptionally upregulated in response to starvation and mostly existed as a protein bound to membrane under both growing and starvation conditions. Immunofluorescence microscopy revealed that the intracellular localization of Apg8p changed drastically after shift to starvation. Apg8p resided on unidentified tiny dot structures dispersed in the cytoplasm at growing phase. During starvation, it was localized on large punctate structures, some of which were confirmed to be autophagosomes and autophagic bodies by immuno-EM. Besides these structures, we found that Apg8p was enriched on isolation membranes and in electron less-dense regions, which should contain Apg8p-localized membrane- or lipid-containing structures. These structures would represent intermediate structures during autophagosome formation. Here, we also showed that microtubule does not play an essential role in the autophagy in yeast. The result does not match with the previously proposed role of Apg8/Aut7p, delivery of autophagosome to the vacuole along microtubule. Moreover, it is revealed that autophagosome formation is severely impaired in the apg8 null mutant. Apg8p would play an important role in the autophagosome formation.
Figures
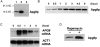
Similar articles
-
Apg2p functions in autophagosome formation on the perivacuolar structure.J Biol Chem. 2001 Aug 10;276(32):30452-60. doi: 10.1074/jbc.M102346200. Epub 2001 May 29. J Biol Chem. 2001. PMID: 11382761
-
The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation.EMBO J. 2001 Nov 1;20(21):5971-81. doi: 10.1093/emboj/20.21.5971. EMBO J. 2001. PMID: 11689437 Free PMC article.
-
Aut2p and Aut7p, two novel microtubule-associated proteins are essential for delivery of autophagic vesicles to the vacuole.EMBO J. 1998 Jul 1;17(13):3597-607. doi: 10.1093/emboj/17.13.3597. EMBO J. 1998. PMID: 9649430 Free PMC article.
-
The yeast Saccharomyces cerevisiae: an overview of methods to study autophagy progression.Methods. 2015 Mar;75:3-12. doi: 10.1016/j.ymeth.2014.12.008. Epub 2014 Dec 16. Methods. 2015. PMID: 25526918 Free PMC article. Review.
-
[Characterization of yeast Atg proteins].Tanpakushitsu Kakusan Koso. 2006 Aug;51(10 Suppl):1457-63. Tanpakushitsu Kakusan Koso. 2006. PMID: 16922419 Review. Japanese. No abstract available.
Cited by
-
Effector Protein Cig2 Decreases Host Tolerance of Infection by Directing Constitutive Fusion of Autophagosomes with the Coxiella-Containing Vacuole.mBio. 2016 Jul 19;7(4):e01127-16. doi: 10.1128/mBio.01127-16. mBio. 2016. PMID: 27435465 Free PMC article.
-
Role of Autophagy in Male Reproductive Processes in Land Plants.Front Plant Sci. 2020 Jun 17;11:756. doi: 10.3389/fpls.2020.00756. eCollection 2020. Front Plant Sci. 2020. PMID: 32625219 Free PMC article. Review.
-
Mcl-1 is a key regulator of the ovarian reserve.Cell Death Dis. 2015 May 7;6(5):e1755. doi: 10.1038/cddis.2015.95. Cell Death Dis. 2015. PMID: 25950485 Free PMC article.
-
Structure of the Atg12-Atg5 conjugate reveals a platform for stimulating Atg8-PE conjugation.EMBO Rep. 2013 Feb;14(2):206-11. doi: 10.1038/embor.2012.208. Epub 2012 Dec 14. EMBO Rep. 2013. PMID: 23238393 Free PMC article.
-
Accessory Interaction Motifs in the Atg19 Cargo Receptor Enable Strong Binding to the Clustered Ubiquitin-related Atg8 Protein.J Biol Chem. 2016 Sep 2;291(36):18799-808. doi: 10.1074/jbc.M116.736892. Epub 2016 Jul 11. J Biol Chem. 2016. PMID: 27402840 Free PMC article.
References
-
- Baba M., Osumi M., Ohsumi Y. Analysis of the membrane structures involved in autophagy in yeast by freeze-replica method. Cell Struct. Funct. 1995;20:465–471. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

