Disruption of the murine nuclear factor I-A gene (Nfia) results in perinatal lethality, hydrocephalus, and agenesis of the corpus callosum
- PMID: 10518556
- PMCID: PMC18392
- DOI: 10.1073/pnas.96.21.11946
Disruption of the murine nuclear factor I-A gene (Nfia) results in perinatal lethality, hydrocephalus, and agenesis of the corpus callosum
Erratum in
- Proc Natl Acad Sci U S A 2001 Mar 27;98(7):4276. Godinho F [corrected to Tolentino-Silva F]
Abstract
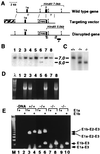
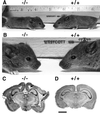
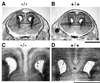
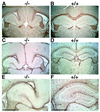
The phylogenetically conserved nuclear factor I (NFI) family of transcription/replication proteins is essential both for adenoviral DNA replication and for the transcription of many cellular genes. We showed previously that the four murine NFI genes (Nfia, Nfib, Nfic, and Nfix) are expressed in unique but overlapping patterns during mouse development and in adult tissues. Here we show that disruption of the Nfia gene causes perinatal lethality, with >95% of homozygous Nfia(-/-) animals dying within 2 weeks after birth. Newborn Nfia(-/-) animals lack a corpus callosum and show ventricular dilation indicating early hydrocephalus. Rare surviving homozygous Nfia(-/-) mice lack a corpus callosum, show severe communicating hydrocephalus, a full-axial tremor indicative of neurological defects, male-sterility, low female fertility, but near normal life spans. These findings indicate that while the Nfia gene appears nonessential for cell viability and DNA replication in embryonic stem cells and fibroblasts, loss of Nfia function causes severe developmental defects. This finding of an NFI gene required for a developmental process suggests that the four NFI genes may have distinct roles in vertebrate development.
Figures
Similar articles
-
Abnormal development of forebrain midline glia and commissural projections in Nfia knock-out mice.J Neurosci. 2003 Jan 1;23(1):203-12. doi: 10.1523/JNEUROSCI.23-01-00203.2003. J Neurosci. 2003. PMID: 12514217 Free PMC article.
-
Expression patterns of the four nuclear factor I genes during mouse embryogenesis indicate a potential role in development.Dev Dyn. 1997 Mar;208(3):313-25. doi: 10.1002/(SICI)1097-0177(199703)208:3<313::AID-AJA3>3.0.CO;2-L. Dev Dyn. 1997. PMID: 9056636
-
Nuclear factor I X deficiency causes brain malformation and severe skeletal defects.Mol Cell Biol. 2007 May;27(10):3855-3867. doi: 10.1128/MCB.02293-06. Mol Cell Biol. 2007. PMID: 17353270 Free PMC article.
-
Roles of the NFI/CTF gene family in transcription and development.Gene. 2000 May 16;249(1-2):31-45. doi: 10.1016/s0378-1119(00)00140-2. Gene. 2000. PMID: 10831836 Review.
-
Variants in nuclear factor I genes influence growth and development.Am J Med Genet C Semin Med Genet. 2019 Dec;181(4):611-626. doi: 10.1002/ajmg.c.31747. Epub 2019 Nov 15. Am J Med Genet C Semin Med Genet. 2019. PMID: 31730271 Review.
Cited by
-
Sensory and spinal inhibitory dorsal midline crossing is independent of Robo3.Front Neural Circuits. 2015 Jul 23;9:36. doi: 10.3389/fncir.2015.00036. eCollection 2015. Front Neural Circuits. 2015. PMID: 26257608 Free PMC article.
-
Astrocytes and disease: a neurodevelopmental perspective.Genes Dev. 2012 May 1;26(9):891-907. doi: 10.1101/gad.188326.112. Genes Dev. 2012. PMID: 22549954 Free PMC article. Review.
-
Specific glial populations regulate hippocampal morphogenesis.J Neurosci. 2008 Nov 19;28(47):12328-40. doi: 10.1523/JNEUROSCI.4000-08.2008. J Neurosci. 2008. PMID: 19020026 Free PMC article.
-
Region-Specific Transcriptional Control of Astrocyte Function Oversees Local Circuit Activities.Neuron. 2020 Jun 17;106(6):992-1008.e9. doi: 10.1016/j.neuron.2020.03.025. Epub 2020 Apr 21. Neuron. 2020. PMID: 32320644 Free PMC article.
-
DNA-binding specificity and in vivo targets of Caenorhabditis elegans nuclear factor I.Proc Natl Acad Sci U S A. 2009 Jul 21;106(29):12049-54. doi: 10.1073/pnas.0812894106. Epub 2009 Jul 7. Proc Natl Acad Sci U S A. 2009. PMID: 19584245 Free PMC article.
References
-
- Hurwitz J, Adhya S, Field J, Gronostajski R, Guggenheimer R A, Kenny M, Lindenbaum J, Nagata K. In: Synthesis of Adenoviral DNA with Purified Proteins. Marks P A, editor. New York: Academic; 1985. pp. 15–24.
-
- Hay R T. J Mol Biol. 1985;186:129–136. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

