Identification of Mycobacterium tuberculosis RNAs synthesized in response to phagocytosis by human macrophages by selective capture of transcribed sequences (SCOTS)
- PMID: 10500215
- PMCID: PMC18072
- DOI: 10.1073/pnas.96.20.11554
Identification of Mycobacterium tuberculosis RNAs synthesized in response to phagocytosis by human macrophages by selective capture of transcribed sequences (SCOTS)
Abstract
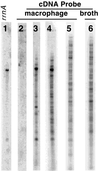
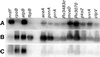
A widely applicable, positive cDNA selection method was developed to identify RNAs synthesized by Mycobacterium tuberculosis in response to phagocytosis by cultured human primary macrophages. cDNAs for sigE and sigH (alternative sigma factors), aceA (isocitrate lyase), ponA (class I penicillin-binding protein), pks2 (polyketide synthase), uvrA (UvrABC endonuclease), and ctpV (putative cation transporter) were obtained from macrophage-grown bacteria. cDNAs for ORFs Rv3070, Rv3483c, Rv0903c (encoding a putative bacterial two-component transcriptional activator), and Rv0170 of the mce1 virulence operon also were obtained from phagocytized bacilli. cDNAs for these genomic regions were not obtained from approximately 1, 000-fold more bacteria grown in laboratory broth. Methods described here, which have identified M. tuberculosis genes expressed in response to host interaction, will allow the study of gene expression in a variety of microorganisms, including expression resulting from interaction with human tissues in natural disease states.
Figures



Similar articles
-
Mycobacterium tuberculosis H37Rv comparative gene-expression analysis in synthetic medium and human macrophage.Gene. 2000 Aug 8;253(2):281-91. doi: 10.1016/s0378-1119(00)00249-3. Gene. 2000. PMID: 10940566
-
Mycobacterium tuberculosis sigma factor E regulon modulates the host inflammatory response.J Infect Dis. 2008 Sep 15;198(6):877-85. doi: 10.1086/591098. J Infect Dis. 2008. PMID: 18657035
-
Global expression analysis of two-component system regulator genes during Mycobacterium tuberculosis growth in human macrophages.FEMS Microbiol Lett. 2004 Jul 15;236(2):341-7. doi: 10.1016/j.femsle.2004.06.010. FEMS Microbiol Lett. 2004. PMID: 15251217
-
Extra and intracellular expression of Mycobacterium tuberculosis genes.Tuber Lung Dis. 1998;79(2):91-7. doi: 10.1054/tuld.1998.0010. Tuber Lung Dis. 1998. PMID: 10645446 Review.
-
The sigma factors of Mycobacterium tuberculosis.FEMS Microbiol Rev. 2006 Nov;30(6):926-41. doi: 10.1111/j.1574-6976.2006.00040.x. FEMS Microbiol Rev. 2006. PMID: 17064287 Review.
Cited by
-
Competitive metagenomic DNA hybridization identifies host-specific microbial genetic markers in cow fecal samples.Appl Environ Microbiol. 2006 Jun;72(6):4054-60. doi: 10.1128/AEM.00023-06. Appl Environ Microbiol. 2006. PMID: 16751515 Free PMC article.
-
Construction and characterization of a Mycobacterium tuberculosis mutant lacking the alternate sigma factor gene, sigF.Infect Immun. 2000 Oct;68(10):5575-80. doi: 10.1128/IAI.68.10.5575-5580.2000. Infect Immun. 2000. PMID: 10992456 Free PMC article.
-
Identification of novel virulence-associated genes via genome analysis of hypothetical genes.Infect Immun. 2004 Mar;72(3):1333-40. doi: 10.1128/IAI.72.3.1333-1340.2004. Infect Immun. 2004. PMID: 14977936 Free PMC article.
-
The extracytoplasmic sigma factor, final sigma(E), is required for intracellular survival of nontypeable Haemophilus influenzae in J774 macrophages.Infect Immun. 2002 Feb;70(2):708-15. doi: 10.1128/IAI.70.2.708-715.2002. Infect Immun. 2002. PMID: 11796603 Free PMC article.
-
Selective capture of Salmonella enterica serovar typhi genes expressed in macrophages that are absent from the Salmonella enterica serovar Typhimurium genome.Infect Immun. 2005 Aug;73(8):5217-21. doi: 10.1128/IAI.73.8.5217-5221.2005. Infect Immun. 2005. PMID: 16041043 Free PMC article.
References
-
- Dannenberg A M, Rook G A W. In: Tuberculosis: Pathogenesis, Protection, and Control. Bloom B R, editor. Washington, DC: ASM Press; 1994. pp. 459–484.
-
- Scott-Craig J S, Guerinot M L, Chelm B K. Mol Gen Genet. 1991;228:356–360. - PubMed
-
- Mahan M J, Slauch J M, Mekalanos J J. Science. 1993;259:686–688. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

