Messenger RNA translation state: the second dimension of high-throughput expression screening
- PMID: 10485877
- PMCID: PMC17934
- DOI: 10.1073/pnas.96.19.10632
Messenger RNA translation state: the second dimension of high-throughput expression screening
Abstract
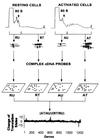
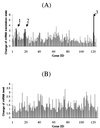
Technological advances over the past 10 years have generated powerful tools for parallel analysis of complex biological problems. Among these new technologies, DNA arrays have provided an important experimental approach for identifying changes in the levels of individual mRNA molecules during important cellular transitions. However, cellular behavior is dictated not by mRNA levels, but by the proteins translated from the individual mRNA species. We report a high-throughput method for simultaneously monitoring the translation state and level of individual mRNA species. Messenger RNAs from resting and mitogenically activated fibroblasts were separated, according to degree of ribosome loading, into well-translated and under-translated pools. cDNA probes generated from these fractions were used to interrogate cDNA arrays. Among approximately 1,200 genes analyzed, less than 1% were found to be translationally regulated in response to mitogenic activation, demonstrating the strong selectivity of this regulatory mechanism. This high-throughput approach is shown to be an effective tool for superimposing translation profile on mRNA level for large numbers of genes, as well as for identifying translationally regulated genes for further study.
Figures


Similar articles
-
Isolation of translationally controlled mRNAs by differential screening.FASEB J. 2000 Aug;14(11):1641-52. doi: 10.1096/fj.14.11.1641. FASEB J. 2000. PMID: 10928999
-
Identifying translationally regulated genes during stem cell differentiation.Curr Protoc Stem Cell Biol. 2011 Sep;Chapter 1:Unit1B.8. doi: 10.1002/9780470151808.sc01b08s18. Curr Protoc Stem Cell Biol. 2011. PMID: 21913168
-
Global mRNA stabilization preferentially linked to translational repression during the endoplasmic reticulum stress response.Mol Cell Biol. 2004 Aug;24(15):6773-87. doi: 10.1128/MCB.24.15.6773-6787.2004. Mol Cell Biol. 2004. PMID: 15254244 Free PMC article.
-
Genome-wide analysis of mRNA polysomal profiles with spotted DNA microarrays.Methods Enzymol. 2007;431:177-201. doi: 10.1016/S0076-6879(07)31010-0. Methods Enzymol. 2007. PMID: 17923236 Review.
-
[Transcriptomes for serial analysis of gene expression].J Soc Biol. 2002;196(4):303-7. J Soc Biol. 2002. PMID: 12645300 Review. French.
Cited by
-
Toward a genome-wide landscape of translational control.Cold Spring Harb Perspect Biol. 2013 Jan 1;5(1):a012302. doi: 10.1101/cshperspect.a012302. Cold Spring Harb Perspect Biol. 2013. PMID: 23209130 Free PMC article. Review.
-
Translational control in endothelial cells.J Vasc Surg. 2007 Jun;45 Suppl A(Suppl A):A8-14. doi: 10.1016/j.jvs.2007.02.033. J Vasc Surg. 2007. PMID: 17544019 Free PMC article. Review.
-
One-step affinity purification of the yeast ribosome and its associated proteins and mRNAs.RNA. 2002 Jul;8(7):948-58. doi: 10.1017/s1355838202026018. RNA. 2002. PMID: 12166649 Free PMC article.
-
Immunopurification of polyribosomal complexes of Arabidopsis for global analysis of gene expression.Plant Physiol. 2005 Jun;138(2):624-35. doi: 10.1104/pp.105.059477. Plant Physiol. 2005. PMID: 15955926 Free PMC article.
-
Spotlight on post-transcriptional control in the circadian system.Cell Mol Life Sci. 2011 Jan;68(1):71-83. doi: 10.1007/s00018-010-0513-5. Epub 2010 Aug 30. Cell Mol Life Sci. 2011. PMID: 20803230 Free PMC article. Review.
References
-
- DeRisi J, Penland L, Brown P O, Bittner M L, Meltzer P S, Ray M, Chen Y, Su Y A, Trent J M. Nat Genet. 1996;14:457–460. - PubMed
-
- Zhang L, Zhou W, Velculescu V E, Kern S E, Hruban R H, Hamilton S R, Vogelstein B, Kinzler K W. Science. 1997;276:1268–1272. - PubMed
-
- Chee M, Yang R, Hubbell E, Berno A, Huang X C, Stern D, Winkler J, Lockhart D J, Morris M S, Fodor S P. Science. 1996;274:610–614. - PubMed
-
- DeRisi J L, Iyer V R, Brown P O. Science. 1997;278:680–686. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

