Cloning of the human cytomegalovirus (HCMV) genome as an infectious bacterial artificial chromosome in Escherichia coli: a new approach for construction of HCMV mutants
- PMID: 10482582
- PMCID: PMC112849
- DOI: 10.1128/JVI.73.10.8320-8329.1999
Cloning of the human cytomegalovirus (HCMV) genome as an infectious bacterial artificial chromosome in Escherichia coli: a new approach for construction of HCMV mutants
Abstract
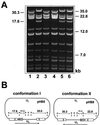
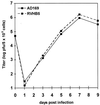
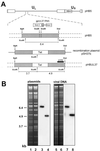
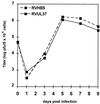
We have recently introduced a novel procedure for the construction of herpesvirus mutants that is based on the cloning and mutagenesis of herpesvirus genomes as infectious bacterial artificial chromosomes (BACs) in Escherichia coli (M. Messerle, I. Crnkovic, W. Hammerschmidt, H. Ziegler, and U. H. Koszinowski, Proc. Natl. Acad. Sci. USA 94:14759-14763, 1997). Here we describe the application of this technique to the human cytomegalovirus (HCMV) strain AD169. Since it was not clear whether the terminal and internal repeat sequences of the HCMV genome would give rise to recombination, the stability of the cloned HCMV genome was examined during propagation in E. coli, during mutagenesis, and after transfection in permissive fibroblasts. Interestingly, the HCMV BACs were frozen in defined conformations in E. coli. The transfection of the HCMV BACs into human fibroblasts resulted in the reconstitution of infectious virus and isomerization of the reconstituted genomes. The power of the BAC mutagenesis procedure was exemplarily demonstrated by the disruption of the gpUL37 open reading frame. The transfection of the mutated BAC led to plaque formation, indicating that the gpUL37 gene product is dispensable for growth of HCMV in fibroblasts. The new procedure will considerably speed up the construction of HCMV mutants and facilitate genetic analysis of HCMV functions.
Figures


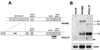
Similar articles
-
Systematic excision of vector sequences from the BAC-cloned herpesvirus genome during virus reconstitution.J Virol. 1999 Aug;73(8):7056-60. doi: 10.1128/JVI.73.8.7056-7060.1999. J Virol. 1999. PMID: 10400809 Free PMC article.
-
Construction of a self-excisable bacterial artificial chromosome containing the human cytomegalovirus genome and mutagenesis of the diploid TRL/IRL13 gene.J Virol. 2002 Mar;76(5):2316-28. doi: 10.1128/jvi.76.5.2316-2328.2002. J Virol. 2002. PMID: 11836410 Free PMC article.
-
Fast screening procedures for random transposon libraries of cloned herpesvirus genomes: mutational analysis of human cytomegalovirus envelope glycoprotein genes.J Virol. 2000 Sep;74(17):7720-9. doi: 10.1128/jvi.74.17.7720-7729.2000. J Virol. 2000. PMID: 10933677 Free PMC article.
-
Cloning of herpesviral genomes as bacterial artificial chromosomes.Rev Med Virol. 2003 Mar-Apr;13(2):111-21. doi: 10.1002/rmv.380. Rev Med Virol. 2003. PMID: 12627394 Review.
-
[BAC system: A novel method for manipulation of herpesvirus genomes based on bacterial genetics].Uirusu. 2004 Dec;54(2):255-64. doi: 10.2222/jsv.54.255. Uirusu. 2004. PMID: 15745165 Review. Japanese.
Cited by
-
Human cytomegalovirus: taking the strain.Med Microbiol Immunol. 2015 Jun;204(3):273-84. doi: 10.1007/s00430-015-0411-4. Epub 2015 Apr 17. Med Microbiol Immunol. 2015. PMID: 25894764 Free PMC article. Review.
-
A human cytomegalovirus gO-null mutant fails to incorporate gH/gL into the virion envelope and is unable to enter fibroblasts and epithelial and endothelial cells.J Virol. 2010 Mar;84(5):2585-96. doi: 10.1128/JVI.02249-09. Epub 2009 Dec 23. J Virol. 2010. PMID: 20032184 Free PMC article.
-
The essential human cytomegalovirus gene UL52 is required for cleavage-packaging of the viral genome.J Virol. 2008 Mar;82(5):2065-78. doi: 10.1128/JVI.01967-07. Epub 2007 Dec 12. J Virol. 2008. PMID: 18077717 Free PMC article.
-
Human cytomegalovirus early protein pUL21a promotes efficient viral DNA synthesis and the late accumulation of immediate-early transcripts.J Virol. 2011 Jan;85(2):663-74. doi: 10.1128/JVI.01599-10. Epub 2010 Nov 3. J Virol. 2011. PMID: 21047969 Free PMC article.
-
Identification of Amino Acids Essential for Viral Replication in the HCMV Helicase-Primase Complex.Front Microbiol. 2018 Oct 23;9:2483. doi: 10.3389/fmicb.2018.02483. eCollection 2018. Front Microbiol. 2018. PMID: 30405556 Free PMC article.
References
-
- Borst, E., G. Pósfai, M. Wagner, and M. Messerle. Unpublished data.
-
- Britt W J, Alford C A. Cytomegalovirus. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2493–2523.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

