A novel Ras-interacting protein required for chemotaxis and cyclic adenosine monophosphate signal relay in Dictyostelium
- PMID: 10473630
- PMCID: PMC25521
- DOI: 10.1091/mbc.10.9.2829
A novel Ras-interacting protein required for chemotaxis and cyclic adenosine monophosphate signal relay in Dictyostelium
Abstract
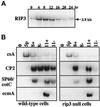
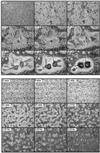
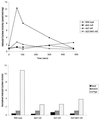
We have identified a novel Ras-interacting protein from Dictyostelium, RIP3, whose function is required for both chemotaxis and the synthesis and relay of the cyclic AMP (cAMP) chemoattractant signal. rip3 null cells are unable to aggregate and lack receptor activation of adenylyl cyclase but are able, in response to cAMP, to induce aggregation-stage, postaggregative, and cell-type-specific gene expression in suspension culture. In addition, rip3 null cells are unable to properly polarize in a cAMP gradient and chemotaxis is highly impaired. We demonstrate that cAMP stimulation of guanylyl cyclase, which is required for chemotaxis, is reduced approximately 60% in rip3 null cells. This reduced activation of guanylyl cyclase may account, in part, for the defect in chemotaxis. When cells are pulsed with cAMP for 5 h to mimic the endogenous cAMP oscillations that occur in wild-type strains, the cells will form aggregates, most of which, however, arrest at the mound stage. Unlike the response seen in wild-type strains, the rip3 null cell aggregates that form under these experimental conditions are very small, which is probably due to the rip3 null cell chemotaxis defect. Many of the phenotypes of the rip3 null cell, including the inability to activate adenylyl cyclase in response to cAMP and defects in chemotaxis, are very similar to those of strains carrying a disruption of the gene encoding the putative Ras exchange factor AleA. We demonstrate that aleA null cells also exhibit a defect in cAMP-mediated activation of guanylyl cyclase similar to that of rip3 null cells. A double-knockout mutant (rip3/aleA null cells) exhibits a further reduction in receptor activation of guanylyl cyclase, and these cells display almost no cell polarization or movement in cAMP gradients. As RIP3 preferentially interacts with an activated form of the Dictyostelium Ras protein RasG, which itself is important for cell movement, we propose that RIP3 and AleA are components of a Ras-regulated pathway involved in integrating chemotaxis and signal relay pathways that are essential for aggregation.
Figures









Similar articles
-
The Dictyostelium MAP kinase kinase DdMEK1 regulates chemotaxis and is essential for chemoattractant-mediated activation of guanylyl cyclase.EMBO J. 1997 Jul 16;16(14):4317-32. doi: 10.1093/emboj/16.14.4317. EMBO J. 1997. PMID: 9250676 Free PMC article.
-
A novel Dictyostelium RasGEF required for chemotaxis and development.BMC Cell Biol. 2005 Dec 7;6:43. doi: 10.1186/1471-2121-6-43. BMC Cell Biol. 2005. PMID: 16336640 Free PMC article.
-
Role of cAMP-dependent protein kinase in controlling aggregation and postaggregative development in Dictyostelium.Dev Biol. 1997 Mar 15;183(2):208-21. doi: 10.1006/dbio.1996.8499. Dev Biol. 1997. PMID: 9126295
-
Transduction of the chemotactic cAMP signal across the plasma membrane of Dictyostelium cells.Experientia. 1995 Dec 18;51(12):1144-54. doi: 10.1007/BF01944732. Experientia. 1995. PMID: 8536802 Review.
-
Novel sensing mechanisms and targets for the cAMP-protein kinase A pathway in the yeast Saccharomyces cerevisiae.Mol Microbiol. 1999 Sep;33(5):904-18. doi: 10.1046/j.1365-2958.1999.01538.x. Mol Microbiol. 1999. PMID: 10476026 Review.
Cited by
-
Involvement of the cytoskeleton in controlling leading-edge function during chemotaxis.Mol Biol Cell. 2010 Jun 1;21(11):1810-24. doi: 10.1091/mbc.e10-01-0009. Epub 2010 Apr 7. Mol Biol Cell. 2010. PMID: 20375144 Free PMC article.
-
PIP3-independent activation of TorC2 and PKB at the cell's leading edge mediates chemotaxis.Curr Biol. 2008 Jul 22;18(14):1034-43. doi: 10.1016/j.cub.2008.06.068. Curr Biol. 2008. PMID: 18635356 Free PMC article.
-
The regulation of cell motility and chemotaxis by phospholipid signaling.J Cell Sci. 2008 Mar 1;121(Pt 5):551-9. doi: 10.1242/jcs.023333. J Cell Sci. 2008. PMID: 18287584 Free PMC article. Review.
-
The mTORC2 signaling network: targets and cross-talks.Biochem J. 2024 Jan 25;481(2):45-91. doi: 10.1042/BCJ20220325. Biochem J. 2024. PMID: 38270460 Free PMC article. Review.
-
Localized Ras signaling at the leading edge regulates PI3K, cell polarity, and directional cell movement.J Cell Biol. 2004 Nov 8;167(3):505-18. doi: 10.1083/jcb.200406177. J Cell Biol. 2004. PMID: 15534002 Free PMC article.
References
-
- Aubry L, Maeda M, Insall R, Devreotes PN, Firtel RA. The Dictyostelium mitogen-activated protein kinase ERK2 is regulated by Ras and cAMP-dependent protein kinase (PKA) and mediates PKA function. J Biol Chem. 1997;272:3883–3886. - PubMed
-
- Burki E, Anjard C, Scholder JC, Reymond CD. Isolation of two genes encoding putative protein kinases regulated during Dictyostelium discoideum development. Gene. 1991;102:57–65. - PubMed
-
- Chen MY, Insall RH, Devreotes PN. Signaling through chemoattractant receptors in Dictyostelium. Trends Genet. 1996;12:52–57. - PubMed
-
- Chung CY, Firtel RA. Dictyostelium: a model experimental system for elucidating the pathways and mechanisms controlling chemotaxis. In: Conn PM, Means A, editors. Molecular Regulation. Totowa, NJ: Humana Press; 1999. (in press).
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

