Systematic changes in gene expression patterns following adaptive evolution in yeast
- PMID: 10449761
- PMCID: PMC22277
- DOI: 10.1073/pnas.96.17.9721
Systematic changes in gene expression patterns following adaptive evolution in yeast
Abstract
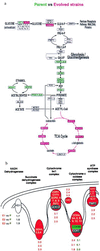
Culturing a population of Saccharomyces cerevisiae for many generations under conditions to which it is not optimally adapted selects for fitter genetic variants. This simple experimental design provides a tractable model of adaptive evolution under natural selection. Beginning with a clonal, founding population, independently evolved strains were obtained from three independent cultures after continuous aerobic growth in glucose-limited chemostats for more than 250 generations. DNA microarrays were used to compare genome-wide patterns of gene expression in the evolved strains and the parental strain. Several hundred genes were found to have significantly altered expression in the evolved strains. Many of these genes showed similar alterations in their expression in all three evolved strains. Genes with altered expression in the three evolved strains included genes involved in glycolysis, the tricarboxylic acid cycle, oxidative phosphorylation, and metabolite transport. These results are consistent with physiological observations and indicate that increased fitness is acquired by altering regulation of central metabolism such that less glucose is fermented and more glucose is completely oxidized.
Figures


Similar articles
-
Genomic analysis of Saccharomyces cerevisiae isolates that grow optimally with glucose as the sole carbon source.Electrophoresis. 2012 Dec;33(23):3514-20. doi: 10.1002/elps.201200172. Epub 2012 Nov 8. Electrophoresis. 2012. PMID: 23135695 Free PMC article.
-
Prolonged selection in aerobic, glucose-limited chemostat cultures of Saccharomyces cerevisiae causes a partial loss of glycolytic capacity.Microbiology (Reading). 2005 May;151(Pt 5):1657-1669. doi: 10.1099/mic.0.27577-0. Microbiology (Reading). 2005. PMID: 15870473
-
Adaptation of Saccharomyces cerevisiae to saline stress through laboratory evolution.J Evol Biol. 2011 May;24(5):1135-53. doi: 10.1111/j.1420-9101.2011.02249.x. Epub 2011 Mar 7. J Evol Biol. 2011. PMID: 21375649
-
Changes in the metabolome of Saccharomyces cerevisiae associated with evolution in aerobic glucose-limited chemostats.FEMS Yeast Res. 2005 Feb;5(4-5):419-30. doi: 10.1016/j.femsyr.2004.11.008. FEMS Yeast Res. 2005. PMID: 15691747
-
The functional basis of adaptive evolution in chemostats.FEMS Microbiol Rev. 2015 Jan;39(1):2-16. doi: 10.1111/1574-6976.12082. Epub 2014 Dec 4. FEMS Microbiol Rev. 2015. PMID: 25098268 Free PMC article. Review.
Cited by
-
Robustness and evolvability in natural chemical resistance: identification of novel systems properties, biochemical mechanisms and regulatory interactions.Mol Biosyst. 2010 Aug;6(8):1475-91. doi: 10.1039/c002567b. Epub 2010 Jun 2. Mol Biosyst. 2010. PMID: 20517567 Free PMC article.
-
High-resolution mutation mapping reveals parallel experimental evolution in yeast.PLoS Biol. 2006 Jul;4(8):e256. doi: 10.1371/journal.pbio.0040256. PLoS Biol. 2006. PMID: 16856782 Free PMC article.
-
Adding confidence to gene expression clustering.Genetics. 2005 Aug;170(4):2003-11. doi: 10.1534/genetics.104.031500. Epub 2005 Jun 8. Genetics. 2005. PMID: 15944369 Free PMC article.
-
Experimental Design, Population Dynamics, and Diversity in Microbial Experimental Evolution.Microbiol Mol Biol Rev. 2018 Jul 25;82(3):e00008-18. doi: 10.1128/MMBR.00008-18. Print 2018 Sep. Microbiol Mol Biol Rev. 2018. PMID: 30045954 Free PMC article. Review.
-
Hap4p overexpression in glucose-grown Saccharomyces cerevisiae induces cells to enter a novel metabolic state.Genome Biol. 2003;4(1):R3. doi: 10.1186/gb-2002-4-1-r3. Epub 2002 Dec 17. Genome Biol. 2003. PMID: 12537548 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

