Mapping of the coronavirus membrane protein domains involved in interaction with the spike protein
- PMID: 10438834
- PMCID: PMC104271
- DOI: 10.1128/JVI.73.9.7441-7452.1999
Mapping of the coronavirus membrane protein domains involved in interaction with the spike protein
Abstract
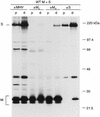
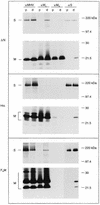
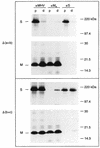
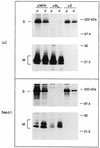
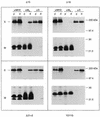
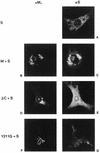
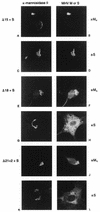
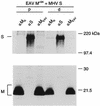
The coronavirus membrane (M) protein is the key player in virion assembly. One of its functions is to mediate the incorporation of the spikes into the viral envelope. Heterotypic interactions between M and the spike (S) protein can be demonstrated by coimmunoprecipitation and by immunofluorescence colocalization, after coexpression of their genes in eukaryotic cells. Using these assays in a mutagenetic approach, we have mapped the domains in the M protein that are involved in complex formation between M and S. It appeared that the 25-residue luminally exposed amino-terminal domain of the M protein is not important for M-S interaction. A 15-residue deletion, the insertion of a His tag, and replacement of the ectodomain by that of another coronavirus M protein did not affect the ability of the M protein to associate with the S protein. However, complex formation was sensitive to changes in the transmembrane domains of this triple-spanning protein. Deletion of either the first two or the last two transmembrane domains, known not to affect the topology of the protein, led to a considerable decrease in complex formation, but association was not completely abrogated. Various effects of changes in the part of the M protein that is located at the cytoplasmic face of the membrane were observed. Deletions of the extreme carboxy-terminal tail appeared not to interfere with M-S complex formation. However, deletions in the amphipathic domain severely affected M-S interaction. Interestingly, changes in the amino-terminal and extreme carboxy-terminal domains of M, which did not disrupt the interaction with S, are known to be fatal to the ability of the protein to engage in virus particle formation (C. A. M. de Haan, L. Kuo, P. S. Masters, H. Vennema, and P. J. M. Rottier, J. Virol. 72:6838-6850, 1998). Apparently, the structural requirements of the M protein for virus particle assembly differ from the requirements for the formation of M-S complexes.
Figures

Similar articles
-
Assembly of the coronavirus envelope: homotypic interactions between the M proteins.J Virol. 2000 Jun;74(11):4967-78. doi: 10.1128/jvi.74.11.4967-4978.2000. J Virol. 2000. PMID: 10799570 Free PMC article.
-
Spike protein assembly into the coronavirion: exploring the limits of its sequence requirements.Virology. 2005 Apr 10;334(2):306-18. doi: 10.1016/j.virol.2005.02.001. Virology. 2005. PMID: 15780881 Free PMC article.
-
Coronavirus envelope assembly is sensitive to changes in the terminal regions of the viral M protein.Adv Exp Med Biol. 1998;440:367-75. doi: 10.1007/978-1-4615-5331-1_48. Adv Exp Med Biol. 1998. PMID: 9782305
-
Receptor specificity and receptor-induced conformational changes in mouse hepatitis virus spike glycoprotein.Adv Exp Med Biol. 2001;494:173-81. doi: 10.1007/978-1-4615-1325-4_29. Adv Exp Med Biol. 2001. PMID: 11774465 Review. No abstract available.
-
Site directed mutagenesis of the murine coronavirus spike protein. Effects on fusion.Adv Exp Med Biol. 1995;380:283-6. doi: 10.1007/978-1-4615-1899-0_45. Adv Exp Med Biol. 1995. PMID: 8830493 Review.
Cited by
-
Sequential glycosylations at the multibasic cleavage site of SARS-CoV-2 spike protein regulate viral activity.Nat Commun. 2024 May 16;15(1):4162. doi: 10.1038/s41467-024-48503-x. Nat Commun. 2024. PMID: 38755139 Free PMC article.
-
Synthesis, insertion, and characterization of SARS-CoV-2 membrane protein within lipid bilayers.Sci Adv. 2024 Mar;10(9):eadm7030. doi: 10.1126/sciadv.adm7030. Epub 2024 Feb 28. Sci Adv. 2024. PMID: 38416838 Free PMC article.
-
Crystal structure of the membrane (M) protein from a bat betacoronavirus.PNAS Nexus. 2023 Jan 30;2(2):pgad021. doi: 10.1093/pnasnexus/pgad021. eCollection 2023 Feb. PNAS Nexus. 2023. PMID: 36874273 Free PMC article.
-
Structure of SARS-CoV-2 M protein in lipid nanodiscs.Elife. 2022 Oct 20;11:e81702. doi: 10.7554/eLife.81702. Elife. 2022. PMID: 36264056 Free PMC article.
-
Characterization and Structural Prediction of Proteins in SARS-CoV-2 Bangladeshi Variant Through Bioinformatics.Microbiol Insights. 2022 Aug 9;15:11786361221115595. doi: 10.1177/11786361221115595. eCollection 2022. Microbiol Insights. 2022. PMID: 35966939 Free PMC article.
References
-
- Brian D A, Hogue B G, Kienzle T E. The coronavirus hemagglutinin esterase glycoprotein. In: Siddell S G, editor. The Coronaviridae. New York, N.Y: Plenum Press; 1995. pp. 165–179.
-
- Cavanagh D. The coronavirus surface glycoprotein. In: Siddell S G, editor. The Coronaviridae. New York, N.Y: Plenum Press; 1995. pp. 73–113.
-
- de Haan C A M, Roestenberg P, de Wit M, de Vries A A F, Nilsson T, Vennema H, Rottier P J M. Structural requirements for O-glycosylation of the mouse hepatitis virus membrane protein. J Biol Chem. 1998;273:29905–29914. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

