A read-ahead function in archaeal DNA polymerases detects promutagenic template-strand uracil
- PMID: 10430892
- PMCID: PMC17729
- DOI: 10.1073/pnas.96.16.9045
A read-ahead function in archaeal DNA polymerases detects promutagenic template-strand uracil
Abstract
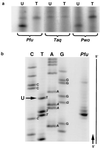
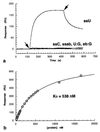
Deamination of cytosine to uracil is the most common promutagenic change in DNA, and it is greatly increased at the elevated growth temperatures of hyperthermophilic archaea. If not repaired to cytosine prior to replication, uracil in a template strand directs incorporation of adenine, generating a G.C --> A.U transition mutation in half the progeny. Surprisingly, genomic analysis of archaea has so far failed to reveal any homologues of either of the known families of uracil-DNA glycosylases responsible for initiating the base-excision repair of uracil in DNA, which is otherwise universal. Here we show that DNA polymerases from several hyperthermophilic archaea (including Vent and Pfu) specifically recognize the presence of uracil in a template strand and stall DNA synthesis before mutagenic misincorporation of adenine. A specific template-checking function in a DNA polymerase has not been observed previously, and it may represent the first step in a pathway for the repair of cytosine deamination in archaea.
Figures



Similar articles
-
Uracil recognition by replicative DNA polymerases is limited to the archaea, not occurring with bacteria and eukarya.Nucleic Acids Res. 2008 Feb;36(3):705-11. doi: 10.1093/nar/gkm1023. Epub 2007 Nov 21. Nucleic Acids Res. 2008. PMID: 18032433 Free PMC article.
-
Structural basis for uracil recognition by archaeal family B DNA polymerases.Nat Struct Biol. 2002 Dec;9(12):922-7. doi: 10.1038/nsb867. Nat Struct Biol. 2002. PMID: 12415291
-
Unwinding of primer-templates by archaeal family-B DNA polymerases in response to template-strand uracil.Nucleic Acids Res. 2013 Feb 1;41(4):2466-78. doi: 10.1093/nar/gks1364. Epub 2013 Jan 8. Nucleic Acids Res. 2013. PMID: 23303790 Free PMC article.
-
Recognition of deaminated bases by archaeal family-B DNA polymerases.Biochem Soc Trans. 2009 Feb;37(Pt 1):65-8. doi: 10.1042/BST0370065. Biochem Soc Trans. 2009. PMID: 19143603 Review.
-
Error-free versus mutagenic processing of genomic uracil--relevance to cancer.DNA Repair (Amst). 2014 Jul;19:38-47. doi: 10.1016/j.dnarep.2014.03.028. Epub 2014 Apr 18. DNA Repair (Amst). 2014. PMID: 24746924 Review.
Cited by
-
Replicative DNA polymerases.Cold Spring Harb Perspect Biol. 2013 Jun 1;5(6):a012799. doi: 10.1101/cshperspect.a012799. Cold Spring Harb Perspect Biol. 2013. PMID: 23732474 Free PMC article. Review.
-
Archaeal replicative primases can perform translesion DNA synthesis.Proc Natl Acad Sci U S A. 2015 Feb 17;112(7):E633-8. doi: 10.1073/pnas.1412982112. Epub 2015 Feb 2. Proc Natl Acad Sci U S A. 2015. PMID: 25646444 Free PMC article.
-
Sulfolobus acidocaldarius UDG Can Remove dU from the RNA Backbone: Insight into the Specific Recognition of Uracil Linked with Deoxyribose.Genes (Basel). 2017 Jan 18;8(1):38. doi: 10.3390/genes8010038. Genes (Basel). 2017. PMID: 28106786 Free PMC article.
-
Comprehensive genetic screening for vascular Ehlers-Danlos syndrome through an amplification-based next-generation sequencing system.Am J Med Genet A. 2023 Jan;191(1):37-51. doi: 10.1002/ajmg.a.62982. Epub 2022 Oct 3. Am J Med Genet A. 2023. PMID: 36189931 Free PMC article.
-
APOBEC3G is a single-stranded DNA cytidine deaminase and functions independently of HIV reverse transcriptase.Nucleic Acids Res. 2004 Apr 30;32(8):2421-9. doi: 10.1093/nar/gkh554. Print 2004. Nucleic Acids Res. 2004. PMID: 15121899 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

