Sec1p binds to SNARE complexes and concentrates at sites of secretion
- PMID: 10427089
- PMCID: PMC3206579
- DOI: 10.1083/jcb.146.2.333
Sec1p binds to SNARE complexes and concentrates at sites of secretion
Abstract
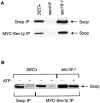
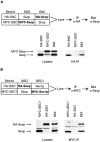
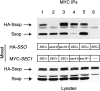
Proteins of the Sec1 family have been shown to interact with target-membrane t-SNAREs that are homologous to the neuronal protein syntaxin. We demonstrate that yeast Sec1p coprecipitates not only the syntaxin homologue Ssop, but also the other two exocytic SNAREs (Sec9p and Sncp) in amounts and in proportions characteristic of SNARE complexes in yeast lysates. The interaction between Sec1p and Ssop is limited by the abundance of SNARE complexes present in sec mutants that are defective in either SNARE complex assembly or disassembly. Furthermore, the localization of green fluorescent protein (GFP)-tagged Sec1p coincides with sites of vesicle docking and fusion where SNARE complexes are believed to assemble and function. The proposal that SNARE complexes act as receptors for Sec1p is supported by the mislocalization of GFP-Sec1p in a mutant defective for SNARE complex assembly and by the robust localization of GFP-Sec1p in a mutant that fails to disassemble SNARE complexes. The results presented here place yeast Sec1p at the core of the exocytic fusion machinery, bound to SNARE complexes and localized to sites of secretion.
Figures

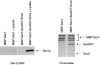
Similar articles
-
Ordering the final events in yeast exocytosis.J Cell Biol. 2000 Oct 16;151(2):439-52. doi: 10.1083/jcb.151.2.439. J Cell Biol. 2000. PMID: 11038189 Free PMC article.
-
Geranylgeranylated SNAREs are dominant inhibitors of membrane fusion.J Cell Biol. 2000 Oct 16;151(2):453-66. doi: 10.1083/jcb.151.2.453. J Cell Biol. 2000. PMID: 11038190 Free PMC article.
-
Molecular interactions position Mso1p, a novel PTB domain homologue, in the interface of the exocyst complex and the exocytic SNARE machinery in yeast.Mol Biol Cell. 2005 Oct;16(10):4543-56. doi: 10.1091/mbc.e05-03-0243. Epub 2005 Jul 19. Mol Biol Cell. 2005. PMID: 16030256 Free PMC article.
-
The role of Sec1p-related proteins in vesicle trafficking in the nerve terminal.J Neurosci Res. 1996 Jul 15;45(2):89-95. doi: 10.1002/(SICI)1097-4547(19960715)45:2<89::AID-JNR1>3.0.CO;2-B. J Neurosci Res. 1996. PMID: 8843026 Review.
-
Synaptic vesicle docking: a putative role for the Munc18/Sec1 protein family.Curr Top Dev Biol. 2005;65:83-113. doi: 10.1016/S0070-2153(04)65003-4. Curr Top Dev Biol. 2005. PMID: 15642380 Review. No abstract available.
Cited by
-
Ultrahigh-resolution imaging reveals formation of neuronal SNARE/Munc18 complexes in situ.Proc Natl Acad Sci U S A. 2013 Jul 23;110(30):E2812-20. doi: 10.1073/pnas.1310654110. Epub 2013 Jul 2. Proc Natl Acad Sci U S A. 2013. PMID: 23821748 Free PMC article.
-
Role of secretory carrier membrane protein SCAMP2 in granule exocytosis.Mol Biol Cell. 2002 Dec;13(12):4266-78. doi: 10.1091/mbc.e02-03-0136. Mol Biol Cell. 2002. PMID: 12475951 Free PMC article.
-
Structure of the Munc18c/Syntaxin4 N-peptide complex defines universal features of the N-peptide binding mode of Sec1/Munc18 proteins.Proc Natl Acad Sci U S A. 2007 May 22;104(21):8773-8. doi: 10.1073/pnas.0701124104. Epub 2007 May 16. Proc Natl Acad Sci U S A. 2007. PMID: 17517664 Free PMC article.
-
The Polarized Redistribution of the Contractile Vacuole to the Rear of the Cell is Critical for Streaming and is Regulated by PI(4,5)P2-Mediated Exocytosis.Front Cell Dev Biol. 2022 Jul 19;9:765316. doi: 10.3389/fcell.2021.765316. eCollection 2021. Front Cell Dev Biol. 2022. PMID: 35928786 Free PMC article. Review.
-
Munc18-1 binding to the neuronal SNARE complex controls synaptic vesicle priming.J Cell Biol. 2009 Mar 9;184(5):751-64. doi: 10.1083/jcb.200812026. Epub 2009 Mar 2. J Cell Biol. 2009. PMID: 19255244 Free PMC article.
References
-
- Abeliovich H., Grote E., Novick P., Ferro-Novick S. Tlg2p, a yeast syntaxin homolog that resides on the Golgi and endocytic structures. J. Biol. Chem. 1998;273:11719–11727. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

