Chs7p, a new protein involved in the control of protein export from the endoplasmic reticulum that is specifically engaged in the regulation of chitin synthesis in Saccharomyces cerevisiae
- PMID: 10366589
- PMCID: PMC2133151
- DOI: 10.1083/jcb.145.6.1153
Chs7p, a new protein involved in the control of protein export from the endoplasmic reticulum that is specifically engaged in the regulation of chitin synthesis in Saccharomyces cerevisiae
Erratum in
- J Cell Biol 1999 Jul 12;146(1):following 264
Abstract
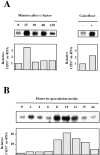
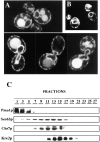
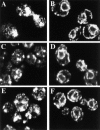
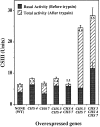
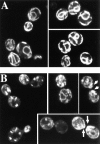
The Saccharomyces cerevisiae CHS7 gene encodes an integral membrane protein located in the ER which is directly involved in chitin synthesis through the regulation of chitin synthase III (CSIII) activity. In the absence of CHS7 product, Chs3p, but not other secreted proteins, is retained in the ER, leading to a severe defect in CSIII activity and consequently, to a reduced rate of chitin synthesis. In addition, chs7 null mutants show the yeast phenotypes associated with a lack of chitin: reduced mating efficiency and lack of the chitosan ascospore layer, clear indications of Chs7p function throughout the S. cerevisiae biological cycle. CHS3 overexpression does not lead to increased levels of CSIII because the Chs3p excess is retained in the ER. However, joint overexpression of CHS3 and CHS7 increases the export of Chs3p from the ER and this is accompanied by a concomitant increase in CSIII activity, indicating that the amount of Chs7p is a limiting factor for CSIII activity. Accordingly, CHS7 transcription is increased when elevated amounts of chitin synthesis are detected. These results show that Chs7p forms part of a new mechanism specifically involved in Chs3p export from the ER and consequently, in the regulation of CSIII activity.
Figures


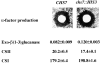
Similar articles
-
Characterization of the chitin biosynthesis process as a compensatory mechanism in the fks1 mutant of Saccharomyces cerevisiae.FEBS Lett. 2000 Jul 28;478(1-2):84-8. doi: 10.1016/s0014-5793(00)01835-4. FEBS Lett. 2000. PMID: 10922474
-
KNR4, a suppressor of Saccharomyces cerevisiae cwh mutants, is involved in the transcriptional control of chitin synthase genes.Microbiology (Reading). 1999 Jan;145 ( Pt 1):249-258. doi: 10.1099/13500872-145-1-249. Microbiology (Reading). 1999. PMID: 10206705
-
Chitin synthase III requires Chs4p-dependent translocation of Chs3p into the plasma membrane.J Cell Sci. 2007 Jun 15;120(Pt 12):1998-2009. doi: 10.1242/jcs.005124. Epub 2007 May 22. J Cell Sci. 2007. PMID: 17519287
-
Chitin Synthesis in Yeast: A Matter of Trafficking.Int J Mol Sci. 2022 Oct 14;23(20):12251. doi: 10.3390/ijms232012251. Int J Mol Sci. 2022. PMID: 36293107 Free PMC article. Review.
-
Biosynthesis of cell wall and septum during yeast growth.Arch Med Res. 1993 Autumn;24(3):301-3. Arch Med Res. 1993. PMID: 8298281 Review.
Cited by
-
Saccharomyces cerevisiae Sps1p regulates trafficking of enzymes required for spore wall synthesis.Eukaryot Cell. 2005 Mar;4(3):536-44. doi: 10.1128/EC.4.3.536-544.2005. Eukaryot Cell. 2005. PMID: 15755916 Free PMC article.
-
The RIM101 pathway contributes to yeast cell wall assembly and its function becomes essential in the absence of mitogen-activated protein kinase Slt2p.Eukaryot Cell. 2006 Mar;5(3):507-17. doi: 10.1128/EC.5.3.507-517.2006. Eukaryot Cell. 2006. PMID: 16524906 Free PMC article.
-
The yeasts Rho1p and Pkc1p regulate the transport of chitin synthase III (Chs3p) from internal stores to the plasma membrane.Proc Natl Acad Sci U S A. 2003 Sep 2;100(18):10287-92. doi: 10.1073/pnas.1834246100. Epub 2003 Aug 19. Proc Natl Acad Sci U S A. 2003. PMID: 12928491 Free PMC article.
-
A novel role of the yeast CaaX protease Ste24 in chitin synthesis.Mol Biol Cell. 2010 Jul 15;21(14):2425-33. doi: 10.1091/mbc.e10-01-0080. Epub 2010 May 26. Mol Biol Cell. 2010. PMID: 20505074 Free PMC article.
-
The exomer coat complex transports Fus1p to the plasma membrane via a novel plasma membrane sorting signal in yeast.Mol Biol Cell. 2009 Dec;20(23):4985-96. doi: 10.1091/mbc.e09-04-0324. Epub 2009 Oct 7. Mol Biol Cell. 2009. PMID: 19812245 Free PMC article.
References
-
- Bulawa CE. Genetics and molecular biology of chitin synthesis in fungi. Annu Rev Microbiol. 1993;47:505–534. - PubMed
-
- Bulawa CE, Slater M, Cabib E, Au-Young J, Sburlati A, Adair WL, Jr, Robbins PW. The S. cerevisiaestructural gene for chitin synthase is not required for chitin synthesis in vivo. Cell. 1986;46:213–225. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

