Subtyping of Haemophilus influenzae strains by pulsed-field gel electrophoresis
- PMID: 10364576
- PMCID: PMC85103
- DOI: 10.1128/JCM.37.7.2142-2147.1999
Subtyping of Haemophilus influenzae strains by pulsed-field gel electrophoresis
Abstract
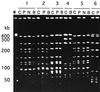
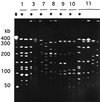
A total of 200 isolates of Haemophilus influenzae were analyzed by serotyping, biotyping, and pulsed-field gel electrophoresis (PFGE). A total of 178 epidemiologically unrelated strains of H. influenzae demonstrated a variety of genome patterns by PFGE, and 165 genotypes were thus obtained in this study. PFGE typing proved to have a much stronger discriminatory power than either serotyping or biotyping. Six serotype b strains were all classified into discrete genotypes. A PFGE analysis of 18 strains obtained from the nasopharynx, blood, and cerebrospinal fluid of patients with meningitis also supported the hypothesis that invasive H. influenzae disseminates from the nasopharynx to the bloodstream and then subsequently to other body sites. PFGE typing of 10 other strains isolated from household contacts of patients with H. influenzae infection revealed that the strain that caused the H. influenzae infection often colonized the nasopharynges of household contacts. Our findings suggest that PFGE analysis is useful for the epidemiological study of H. influenzae infection, even when the invasive disease is caused by serotype b strains.
Figures


Similar articles
-
Invasive serotype a Haemophilus influenzae infections with a virulence genotype resembling Haemophilus influenzae type b: emerging pathogen in the vaccine era?Pediatrics. 2001 Jul;108(1):E18. doi: 10.1542/peds.108.1.e18. Pediatrics. 2001. PMID: 11433097
-
[Genotyping of ampicillin-resistant Haemophilus influenzae].Zhonghua Er Ke Za Zhi. 2005 Sep;43(9):685-9. Zhonghua Er Ke Za Zhi. 2005. PMID: 16191303 Chinese.
-
Use of pulsed-field gel electrophoresis, enterobacterial repetitive intergenic consensus typing, and automated ribotyping to assess genomic variability among strains of nontypeable Haemophilus influenzae.J Clin Microbiol. 2002 Feb;40(2):660-2. doi: 10.1128/JCM.40.2.660-662.2002. J Clin Microbiol. 2002. PMID: 11825990 Free PMC article.
-
Genotyping of Streptococcus pneumoniae and Haemophilus influenzae isolated from paired middle ear fluid and nasopharynx by pulsed-field gel electrophoresis.ORL J Otorhinolaryngol Relat Spec. 2004;66(5):233-40. doi: 10.1159/000081119. ORL J Otorhinolaryngol Relat Spec. 2004. PMID: 15583436
-
Nontypeable Haemophilus influenzae as a pathogen in children.Pediatr Infect Dis J. 2009 Jan;28(1):43-8. doi: 10.1097/INF.0b013e318184dba2. Pediatr Infect Dis J. 2009. PMID: 19057458 Review.
Cited by
-
Haemophilus influenzae: genetic variability and natural selection to identify virulence factors.Infect Immun. 2004 May;72(5):2457-61. doi: 10.1128/IAI.72.5.2457-2461.2004. Infect Immun. 2004. PMID: 15102751 Free PMC article. Review. No abstract available.
-
Limited genetic diversity of recent invasive isolates of non-serotype b encapsulated Haemophilus influenzae.J Clin Microbiol. 2002 Apr;40(4):1264-70. doi: 10.1128/JCM.40.4.1264-1270.2002. J Clin Microbiol. 2002. PMID: 11923343 Free PMC article.
-
Invasive type e Haemophilus influenzae disease in Italy.Emerg Infect Dis. 2003 Feb;9(2):258-61. doi: 10.3201/eid0902.020142. Emerg Infect Dis. 2003. PMID: 12604001 Free PMC article.
-
Antimicrobial resistance in Haemophilus influenzae.Clin Microbiol Rev. 2007 Apr;20(2):368-89. doi: 10.1128/CMR.00040-06. Clin Microbiol Rev. 2007. PMID: 17428889 Free PMC article. Review.
-
Invasive disease due to nontypeable Haemophilus influenzae among children in Arkansas.J Clin Microbiol. 2003 Jul;41(7):3064-9. doi: 10.1128/JCM.41.7.3064-3069.2003. J Clin Microbiol. 2003. PMID: 12843045 Free PMC article.
References
-
- Barenkamp S J, Munson R S J, Granoff D M. Subtyping isolates of Haemophilus influenzae type b by outer-membrane protein profiles. J Infect Dis. 1981;143:668–676. - PubMed
-
- Bol P, Spanjaard L, van Alphen L, Zanen H C. Epidemiology of H. influenzae meningitis in patients more than 6 years of age. J Infect. 1987;15:81–94. - PubMed
-
- Butler P D, Moxon E R. A physical map of the genome of Haemophilus influenzae type b. J Gen Microbiol. 1990;136:2333–2342. - PubMed
-
- Curran R, Hardie K R, Towner K J. Analysis by pulsed-field gel electrophoresis of insertion mutation in the transferrin-binding system of Haemophilus influenzae type b. J Med Microbiol. 1994;41:120–126. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

