Constant mean viral copy number per infected cell in tissues regardless of high, low, or undetectable plasma HIV RNA
- PMID: 10330433
- PMCID: PMC2193638
- DOI: 10.1084/jem.189.10.1545
Constant mean viral copy number per infected cell in tissues regardless of high, low, or undetectable plasma HIV RNA
Abstract
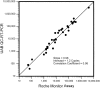
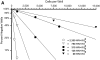
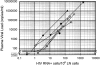
Quantitative analysis of the relationship between virus expression and disease outcome has been critical for understanding HIV-1 pathogenesis. Yet the amount of viral RNA contained within an HIV-expressing cell and the relationship between the number of virus-producing cells and plasma virus load has not been established or reflected in models of viral dynamics. We report here a novel strategy for the coordinated analysis of virus expression in lymph node specimens. The results obtained for patients with a broad range of plasma viral loads before and after antiretroviral therapy reveal a constant mean viral (v)RNA copy number (3.6 log10 copies) per infected cell, regardless of plasma virus load or treatment status. In addition, there was a significant but nonlinear direct correlation between the frequency of vRNA+ lymph node cells and plasma vRNA. As predicted from this relationship, residual cells expressing this same mean copy number are detectable (frequency <2/10(6) cells) in tissues of treated patients who have plasma vRNA levels below the current detectable threshold (<50 copies/ml). These data suggest that fully replication-active cells are responsible for sustaining viremia after initiation of potent antiretroviral therapy and that plasma virus titers correlate, albeit in a nonlinear fashion, with the number of virus-expressing cells in lymphoid tissue.
Figures


Similar articles
-
Residual human immunodeficiency virus (HIV) Type 1 RNA and DNA in lymph nodes and HIV RNA in genital secretions and in cerebrospinal fluid after suppression of viremia for 2 years.J Infect Dis. 2001 May 1;183(9):1318-27. doi: 10.1086/319864. Epub 2001 Apr 10. J Infect Dis. 2001. PMID: 11294662
-
Reduction of HIV-1 in blood and lymph nodes following potent antiretroviral therapy and the virologic correlates of treatment failure.Proc Natl Acad Sci U S A. 1997 Nov 11;94(23):12574-9. doi: 10.1073/pnas.94.23.12574. Proc Natl Acad Sci U S A. 1997. PMID: 9356491 Free PMC article. Clinical Trial.
-
Influence of L-lysine amino acid on the HIV-1 RNA replication in vitro.Antivir Chem Chemother. 2015 Feb;24(1):39-46. doi: 10.1177/2040206614566582. Antivir Chem Chemother. 2015. PMID: 26149265 Free PMC article.
-
Viral load and immunophenotype of cells obtained from lymph nodes by fine needle aspiration as compared with peripheral blood cells in HIV-infected patients.J Acquir Immune Defic Syndr Hum Retrovirol. 1996 Sep;13(1):39-47. doi: 10.1097/00042560-199609000-00007. J Acquir Immune Defic Syndr Hum Retrovirol. 1996. PMID: 8797685
-
Molecular biological assessment methods and understanding the course of the HIV infection.APMIS Suppl. 2003;(114):1-37. APMIS Suppl. 2003. PMID: 14626050 Review.
Cited by
-
Viral and cellular dynamics in HIV disease.Curr HIV/AIDS Rep. 2004 Apr;1(1):40-6. doi: 10.1007/s11904-004-0006-y. Curr HIV/AIDS Rep. 2004. PMID: 16091222 Review.
-
Continued production of drug-sensitive human immunodeficiency virus type 1 in children on combination antiretroviral therapy who have undetectable viral loads.J Virol. 2004 Jan;78(2):968-79. doi: 10.1128/jvi.78.2.968-979.2004. J Virol. 2004. PMID: 14694128 Free PMC article.
-
Residual Viremia in Treated HIV+ Individuals.PLoS Comput Biol. 2016 Jan 6;12(1):e1004677. doi: 10.1371/journal.pcbi.1004677. eCollection 2016 Jan. PLoS Comput Biol. 2016. PMID: 26735135 Free PMC article.
-
Estimating frequencies of minority nevirapine-resistant strains in chronically HIV-1-infected individuals naive to nevirapine by using stochastic simulations and a mathematical model.J Virol. 2010 Oct;84(19):10230-40. doi: 10.1128/JVI.01010-10. Epub 2010 Jul 28. J Virol. 2010. PMID: 20668070 Free PMC article.
-
Modeling latently infected cell activation: viral and latent reservoir persistence, and viral blips in HIV-infected patients on potent therapy.PLoS Comput Biol. 2009 Oct;5(10):e1000533. doi: 10.1371/journal.pcbi.1000533. Epub 2009 Oct 16. PLoS Comput Biol. 2009. PMID: 19834532 Free PMC article.
References
-
- Mellors J, Rinaldo C, Gupta P, White RM, Todd JA, Kingsley LA. Prognosis in HIV infection predicted by the quantity of virus in plasma. Science. 1996;272:1167–1170. - PubMed
-
- O'Brien W, Hartigan P, Martin D. Changes in plasma HIV-1 RNA and CD4+lymphocyte counts and the risk of progression to AIDS. Rapid and simple PCR assay for quantitation of HIV-1 RNA in plasma: application to acute retroviral infection. N Engl J Med. 1996;334:426–431. - PubMed
-
- Carpenter CC, Fischl MA, Hammer SM, Hirsch MS, Jacobsen DM, Katzenstein D, Montaner JS, Richman DD, Saag MS, Schooley RT, et al. Antiretroviral therapy for HIV infection in 1996. Recommendations of an international panel. International AIDS Society-USA. JAMA. 1996;276:146–154. - PubMed
-
- Saag MS, Holodniy M, Kuritzkes DR, O'Brien WA, Coombs R, Poscher ME, Jacobsen DM, Shaw GM, Richman DD, Volberding PA. HIV viral load markers in clinical practice. Nat Med. 1996;2:625–629. - PubMed
-
- Pantaleo G, Graziosi C, Demarest JF, Butini L, Montroni M, Fox CH, Orenstein JM, Kotler DP, Fauci AS. HIV infection is active and progressive in lymphoid tissue during the clinically latent stage of disease. Nature. 1993;362:355–358. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

