Identification of CHIP, a novel tetratricopeptide repeat-containing protein that interacts with heat shock proteins and negatively regulates chaperone functions
- PMID: 10330192
- PMCID: PMC104411
- DOI: 10.1128/MCB.19.6.4535
Identification of CHIP, a novel tetratricopeptide repeat-containing protein that interacts with heat shock proteins and negatively regulates chaperone functions
Abstract
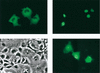
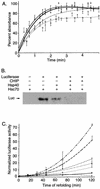
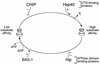
The chaperone function of the mammalian 70-kDa heat shock proteins Hsc70 and Hsp70 is modulated by physical interactions with four previously identified chaperone cofactors: Hsp40, BAG-1, the Hsc70-interacting protein Hip, and the Hsc70-Hsp90-organizing protein Hop. Hip and Hop interact with Hsc70 via a tetratricopeptide repeat domain. In a search for additional tetratricopeptide repeat-containing proteins, we have identified a novel 35-kDa cytoplasmic protein, carboxyl terminus of Hsc70-interacting protein (CHIP). CHIP is highly expressed in adult striated muscle in vivo and is expressed broadly in vitro in tissue culture. Hsc70 and Hsp70 were identified as potential interaction partners for this protein in a yeast two-hybrid screen. In vitro binding assays demonstrated direct interactions between CHIP and both Hsc70 and Hsp70, and complexes containing CHIP and Hsc70 were identified in immunoprecipitates of human skeletal muscle cells in vivo. Using glutathione S-transferase fusions, we found that CHIP interacted with the carboxy-terminal residues 540 to 650 of Hsc70, whereas Hsc70 interacted with the amino-terminal residues 1 to 197 (containing the tetratricopeptide domain and an adjacent charged domain) of CHIP. Recombinant CHIP inhibited Hsp40-stimulated ATPase activity of Hsc70 and Hsp70, suggesting that CHIP blocks the forward reaction of the Hsc70-Hsp70 substrate-binding cycle. Consistent with this observation, both luciferase refolding and substrate binding in the presence of Hsp40 and Hsp70 were inhibited by CHIP. Taken together, these results indicate that CHIP decreases net ATPase activity and reduces chaperone efficiency, and they implicate CHIP in the negative regulation of the forward reaction of the Hsc70-Hsp70 substrate-binding cycle.
Figures


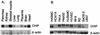



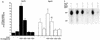
Similar articles
-
Interaction of the Hsp90 cochaperone cyclophilin 40 with Hsc70.Cell Stress Chaperones. 2004 Summer;9(2):167-81. doi: 10.1379/csc-26r.1. Cell Stress Chaperones. 2004. PMID: 15497503 Free PMC article.
-
A brain-specific isoform of small glutamine-rich tetratricopeptide repeat-containing protein binds to Hsc70 and the cysteine string protein.J Biol Chem. 2003 Oct 3;278(40):38376-83. doi: 10.1074/jbc.M301558200. Epub 2003 Jul 23. J Biol Chem. 2003. PMID: 12878599
-
The carboxy-terminal domain of Hsc70 provides binding sites for a distinct set of chaperone cofactors.Mol Cell Biol. 1998 Apr;18(4):2023-8. doi: 10.1128/MCB.18.4.2023. Mol Cell Biol. 1998. PMID: 9528774 Free PMC article.
-
Regulation of the heat shock conjugate Hsc70 in the mammalian cell: the characterization of the anti-apoptotic protein BAG-1 provides novel insights.Biol Chem. 1998 Mar;379(3):269-74. Biol Chem. 1998. PMID: 9563821 Review.
-
CHIP: a link between the chaperone and proteasome systems.Cell Stress Chaperones. 2003 Winter;8(4):303-8. doi: 10.1379/1466-1268(2003)008<0303:calbtc>2.0.co;2. Cell Stress Chaperones. 2003. PMID: 15115282 Free PMC article. Review.
Cited by
-
Post-Translational Regulations of Foxp3 in Treg Cells and Their Therapeutic Applications.Front Immunol. 2021 Apr 12;12:626172. doi: 10.3389/fimmu.2021.626172. eCollection 2021. Front Immunol. 2021. PMID: 33912156 Free PMC article. Review.
-
c-IAP1 binds and processes PCSK9 protein: linking the c-IAP1 in a TNF-α pathway to PCSK9-mediated LDLR degradation pathway.Molecules. 2012 Oct 15;17(10):12086-101. doi: 10.3390/molecules171012086. Molecules. 2012. PMID: 23085658 Free PMC article.
-
Cytosolic protein quality control machinery: Interactions of Hsp70 with a network of co-chaperones and substrates.Exp Biol Med (Maywood). 2021 Jun;246(12):1419-1434. doi: 10.1177/1535370221999812. Epub 2021 Mar 17. Exp Biol Med (Maywood). 2021. PMID: 33730888 Free PMC article. Review.
-
A ubiquitin-specific, proximity-based labeling approach for the identification of ubiquitin ligase substrates.Sci Adv. 2024 Aug 9;10(32):eadp3000. doi: 10.1126/sciadv.adp3000. Epub 2024 Aug 9. Sci Adv. 2024. PMID: 39121224 Free PMC article.
-
CHIP protects against cardiac pressure overload through regulation of AMPK.J Clin Invest. 2013 Aug;123(8):3588-99. doi: 10.1172/JCI69080. Epub 2013 Jul 25. J Clin Invest. 2013. PMID: 23863712 Free PMC article.
References
-
- Angelier N, Moreau N, Rodriquez-Martin M L, Penrad-Mobayed M, Prudhomme C. Does the chaperone heat shock protein Hsp70 play a role in the control of developmental processes? Int J Dev Biol. 1996;40:521–529. - PubMed
-
- Ballinger, C. A., and C. Patterson. Unpublished observations.
-
- Benaroudj N, Fouchaq B, Ladjimi M M. The COOH-terminal peptide binding domain is essential for self-association of the molecular chaperone Hsc70. J Biol Chem. 1997;272:8744–8751. - PubMed
-
- Bercovich B, Stancovski I, Mayer A, Blumenfeld N, Laszlo A, Schwartz A L, Ciechanover A. Ubiquitin-dependent degradation of certain protein substrates in vitro requires the molecular chaperone Hsc70. J Biol Chem. 1997;272:9002–9010. - PubMed
-
- Boice J A, Hightower L E. A mutational study of the peptide-binding domain of Hsc70 guided by secondary structure prediction. J Biol Chem. 1997;272:24825–24831. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
