Identification of the R1 oncogene and its protein product from the rhadinovirus of rhesus monkeys
- PMID: 10233975
- PMCID: PMC112557
- DOI: 10.1128/JVI.73.6.5123-5131.1999
Identification of the R1 oncogene and its protein product from the rhadinovirus of rhesus monkeys
Abstract
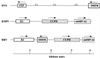
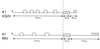
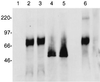
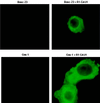
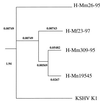
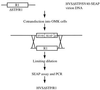
Rhesus monkey rhadinovirus (RRV) is a gamma-2 herpesvirus that is most closely related to the human Kaposi's sarcoma-associated herpesvirus (KSHV). We have identified a distinct open reading frame at the left end of RRV and designated it R1. The position of the R1 gene is equivalent to that of the saimiri transforming protein (STP) of herpesvirus saimiri (HVS) and of K1 of KSHV, other members of the gamma-2 or rhadinovirus subgroup of herpesviruses. The R1 sequence revealed an open reading frame encoding a product of 423 amino acids that was predicted to contain an extracellular domain, a transmembrane domain, and a C-terminal cytoplasmic tail reflective of a type I membrane-bound protein. The predicted structural motifs of R1, including the presence of immunoreceptor tyrosine-based activation motifs, resembled those in K1 of KSHV but were distinct from those of STP. R1 sequences from four independent isolates from three different macaque species revealed 0.95 to 7.3% divergence over the 423 amino acids. Variation was located predominantly within the predicted extracellular domain. The R1 protein migrated at 70 kDa by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and was extensively glycosylated. Tagged R1 protein was localized to the cytoplasmic and plasma membranes of transfected cells. Expression of the R1 gene in Rat-1 fibroblasts induced morphologic changes and focus formation, and injection of R1-expressing cells into nude mice induced the formation of multifocal tumors. A recombinant herpesvirus in which the STP oncogene of HVS was replaced by R1 immortalized T lymphocytes to interleukin-2-independent growth. These results indicate that R1 is an oncogene of RRV.
Figures





Similar articles
-
The primary sequence of rhesus monkey rhadinovirus isolate 26-95: sequence similarities to Kaposi's sarcoma-associated herpesvirus and rhesus monkey rhadinovirus isolate 17577.J Virol. 2000 Apr;74(7):3388-98. doi: 10.1128/jvi.74.7.3388-3398.2000. J Virol. 2000. PMID: 10708456 Free PMC article.
-
Activation of lymphocyte signaling by the R1 protein of rhesus monkey rhadinovirus.J Virol. 2000 Mar;74(6):2721-30. doi: 10.1128/jvi.74.6.2721-2730.2000. J Virol. 2000. PMID: 10684288 Free PMC article.
-
Deregulation of cell growth by the K1 gene of Kaposi's sarcoma-associated herpesvirus.Nat Med. 1998 Apr;4(4):435-40. doi: 10.1038/nm0498-435. Nat Med. 1998. PMID: 9546789
-
Primate herpesviral oncogenes.Mol Cells. 1999 Aug 31;9(4):345-9. Mol Cells. 1999. PMID: 10515596 Review.
-
Regulation of intracellular signalling by the terminal membrane proteins of members of the Gammaherpesvirinae.J Gen Virol. 2006 May;87(Pt 5):1047-1074. doi: 10.1099/vir.0.81598-0. J Gen Virol. 2006. PMID: 16603506 Review.
Cited by
-
Simian homologues of human herpesvirus 8.Philos Trans R Soc Lond B Biol Sci. 2001 Apr 29;356(1408):535-43. doi: 10.1098/rstb.2000.0782. Philos Trans R Soc Lond B Biol Sci. 2001. PMID: 11313010 Free PMC article. Review.
-
Herpesvirus saimiri.Philos Trans R Soc Lond B Biol Sci. 2001 Apr 29;356(1408):545-67. doi: 10.1098/rstb.2000.0780. Philos Trans R Soc Lond B Biol Sci. 2001. PMID: 11313011 Free PMC article. Review.
-
The Modulation of Apoptotic Pathways by Gammaherpesviruses.Front Microbiol. 2016 Apr 27;7:585. doi: 10.3389/fmicb.2016.00585. eCollection 2016. Front Microbiol. 2016. PMID: 27199919 Free PMC article. Review.
-
Evolutionary aspects of oncogenic herpesviruses.Mol Pathol. 2000 Oct;53(5):222-37. doi: 10.1136/mp.53.5.222. Mol Pathol. 2000. PMID: 11091846 Free PMC article. Review.
-
The primary sequence of rhesus monkey rhadinovirus isolate 26-95: sequence similarities to Kaposi's sarcoma-associated herpesvirus and rhesus monkey rhadinovirus isolate 17577.J Virol. 2000 Apr;74(7):3388-98. doi: 10.1128/jvi.74.7.3388-3398.2000. J Virol. 2000. PMID: 10708456 Free PMC article.
References
-
- Boshoff C, Whitby D, Hatziannou T, Fisher C, Van der Walt J, Hatzakis A, Weiss R A, Schultz T F. Kaposi’s sarcoma associated herpesvirus in HIV-negative Kaposi sarcoma. Lancet. 1995;345:1043–1044. - PubMed
-
- Cambier J C. Antigen and Fc receptor signaling. The awesome power of the immunoreceptor tyrosine-based activation motif (ITAM) J Immunol. 1995;155:3281–3285. - PubMed
-
- Cesarman E, Nador R, Knowles D M. Body cavity based lymphoma in an HIV-seronegative patient without Kaposi’s sarcoma associated herpesvirus-like DNA sequences. N Engl J Med. 1996;334:273. - PubMed
-
- Cesarman E, Chang Y, Moore P S, Said J W, Knowles D M. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med. 1995;332:1186–1191. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

