Feline leukemia virus long terminal repeat activates collagenase IV gene expression through AP-1
- PMID: 10233955
- PMCID: PMC112537
- DOI: 10.1128/JVI.73.6.4931-4940.1999
Feline leukemia virus long terminal repeat activates collagenase IV gene expression through AP-1
Abstract
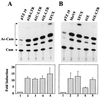
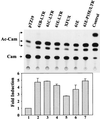
Leukemia and lymphoma induced by feline leukemia viruses (FeLVs) are the commonest forms of illness in domestic cats. These viruses do not contain oncogenes, and the source of their pathogenic activity is not clearly understood. Mechanisms involving proto-oncogene activation subsequent to proviral integration and/or development of recombinant viruses with enhanced replication properties are thought to play an important role in their disease pathogenesis. In addition, the long terminal repeat (LTR) regions of these viruses have been shown to be important determinants for pathogenicity and tissue specificity, by virtue of their ability to interact with various transcription factors. Previously, we have shown that, in the case of Moloney murine leukemia virus, the U3 region of the LTR independently induces transcriptional activation of specific cellular genes through an LTR-generated RNA transcript (S. Y. Choi and D. V. Faller, J. Biol. Chem. 269:19691-19694, 1994; S.-Y. Choi and D. V. Faller, J. Virol. 69:7054-7060, 1995). In this report, we show that the U3 region of exogenous FeLV LTRs can induce transcription from collagenase IV (matrix metalloproteinase 9) and monocyte chemotactic protein 1 (MCP-1) promoters up to 12-fold. We also show that AP-1 DNA-binding activity and transcriptional activity are strongly induced in cells expressing FeLV LTRs and that LTR-specific RNA transcripts are generated in those cells. Activation of mitogen-activated protein kinase kinases 1 and 2 (MEK1 and -2) by the LTR is an intermediate step in the FeLV LTR-mediated induction of AP-1 activity. These findings thus suggest that the LTRs of FeLVs can independently activate transcription of specific cellular genes. This LTR-mediated cellular gene transactivation may play an important role in tumorigenesis or preleukemic states and may be a generalizable activity of leukemia-inducing retroviruses.
Figures


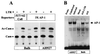

Similar articles
-
Long terminal repeat regions from exogenous but not endogenous feline leukemia viruses transactivate cellular gene expression.J Virol. 2000 Oct;74(20):9742-8. doi: 10.1128/jvi.74.20.9742-9748.2000. J Virol. 2000. PMID: 11000248 Free PMC article.
-
Mutations that abrogate transactivational activity of the feline leukemia virus long terminal repeat do not affect virus replication.Virology. 2003 May 10;309(2):294-305. doi: 10.1016/s0042-6822(03)00069-2. Virology. 2003. PMID: 12758176
-
Leukemia virus long terminal repeat activates NFkappaB pathway by a TLR3-dependent mechanism.Virology. 2006 Feb 20;345(2):390-403. doi: 10.1016/j.virol.2005.10.003. Epub 2005 Nov 14. Virology. 2006. PMID: 16289658 Free PMC article.
-
Advances in understanding molecular determinants in FeLV pathology.Vet Immunol Immunopathol. 2008 May 15;123(1-2):14-22. doi: 10.1016/j.vetimm.2008.01.008. Epub 2008 Jan 19. Vet Immunol Immunopathol. 2008. PMID: 18289704 Free PMC article. Review.
-
Endogenous env elements: partners in generation of pathogenic feline leukemia viruses.Virus Genes. 1995;11(2-3):147-61. doi: 10.1007/BF01728655. Virus Genes. 1995. PMID: 8828142 Review.
Cited by
-
Oncogene cooperativity in Friend erythroleukemia: erythropoietin receptor activation by the env gene of SFFV leads to transcriptional upregulation of PU.1, independent of SFFV proviral insertion.Oncogene. 2002 Feb 14;21(8):1272-84. doi: 10.1038/sj.onc.1205183. Oncogene. 2002. PMID: 11850847 Free PMC article.
-
Antisense transcription in gammaretroviruses as a mechanism of insertional activation of host genes.J Virol. 2010 Apr;84(8):3780-8. doi: 10.1128/JVI.02088-09. Epub 2010 Feb 3. J Virol. 2010. PMID: 20130045 Free PMC article.
-
Molecular detection, phylogenetic analysis, and identification of transcription motifs in feline leukemia virus from naturally infected cats in malaysia.Vet Med Int. 2014;2014:760961. doi: 10.1155/2014/760961. Epub 2014 Nov 17. Vet Med Int. 2014. PMID: 25506469 Free PMC article.
-
Identification of LTR-specific small non-coding RNA in FeLV infected cells.FEBS Lett. 2009 Apr 17;583(8):1386-90. doi: 10.1016/j.febslet.2009.03.056. Epub 2009 Mar 29. FEBS Lett. 2009. PMID: 19336234 Free PMC article.
-
Long terminal repeat regions from exogenous but not endogenous feline leukemia viruses transactivate cellular gene expression.J Virol. 2000 Oct;74(20):9742-8. doi: 10.1128/jvi.74.20.9742-9748.2000. J Virol. 2000. PMID: 11000248 Free PMC article.
References
-
- Alessi D R, Cuenda A, Cohen P, Dudley D T, Saltiel A R. PD 098059 is a specific inhibitor of the activation of mitogen-activated protein kinase kinase in vitro and in vivo. J Biol Chem. 1995;270:27489–27494. - PubMed
-
- Angel P, Hattori K, Smeal T, Karin M. The jun proto-oncogene is positively autoregulated by its product, Jun/AP-1. Cell. 1988;55:875–885. - PubMed
-
- Angel P, Karin M. The role of Jun, Fos and the AP-1 complex in cell-proliferation and transformation. Biochim Biophys Acta. 1991;1072:129–157. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

