Apg7p/Cvt2p: A novel protein-activating enzyme essential for autophagy
- PMID: 10233150
- PMCID: PMC25280
- DOI: 10.1091/mbc.10.5.1367
Apg7p/Cvt2p: A novel protein-activating enzyme essential for autophagy
Abstract
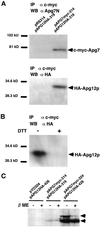
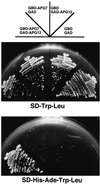
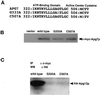
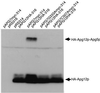
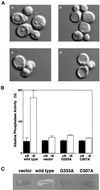
In the yeast Saccharomyces cerevisiae, the Apg12p-Apg5p conjugating system is essential for autophagy. Apg7p is required for the conjugation reaction, because Apg12p is unable to form a conjugate with Apg5p in the apg7/cvt2 mutant. Apg7p shows a significant similarity to a ubiquitin-activating enzyme, Uba1p. In this article, we investigated the function of Apg7p as an Apg12p-activating enzyme. Hemagglutinin-tagged Apg12p was coimmunoprecipitated with c-myc-tagged Apg7p. A two-hybrid experiment confirmed the interaction. The coimmunoprecipitation was sensitive to a thiol-reducing reagent. Furthermore, a thioester conjugate of Apg7p was detected in a lysate of cells overexpressing both Apg7p and Apg12p. These results indicated that Apg12p interacts with Apg7p via a thioester bond. Mutational analyses of Apg7p suggested that Cys507 of Apg7p is an active site cysteine and that both the ATP-binding domain and the cysteine residue are essential for the conjugation of Apg7p with Apg12p to form the Apg12p-Apg5p conjugate. Cells expressing mutant Apg7ps, Apg7pG333A, or Apg7pC507A showed defects in autophagy and cytoplasm-to-vacuole targeting of aminopeptidase I. These results indicated that Apg7p functions as a novel protein-activating enzyme necessary for Apg12p-Apg5p conjugation.
Figures

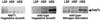
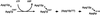
Similar articles
-
The C-terminal region of an Apg7p/Cvt2p is required for homodimerization and is essential for its E1 activity and E1-E2 complex formation.J Biol Chem. 2001 Mar 30;276(13):9846-54. doi: 10.1074/jbc.M007737200. Epub 2001 Jan 3. J Biol Chem. 2001. PMID: 11139573
-
Apg10p, a novel protein-conjugating enzyme essential for autophagy in yeast.EMBO J. 1999 Oct 1;18(19):5234-41. doi: 10.1093/emboj/18.19.5234. EMBO J. 1999. PMID: 10508157 Free PMC article.
-
The human homolog of Saccharomyces cerevisiae Apg7p is a Protein-activating enzyme for multiple substrates including human Apg12p, GATE-16, GABARAP, and MAP-LC3.J Biol Chem. 2001 Jan 19;276(3):1701-6. doi: 10.1074/jbc.C000752200. Epub 2000 Nov 28. J Biol Chem. 2001. PMID: 11096062
-
Molecular mechanism of autophagy in yeast, Saccharomyces cerevisiae.Philos Trans R Soc Lond B Biol Sci. 1999 Sep 29;354(1389):1577-80; discussion 1580-1. doi: 10.1098/rstb.1999.0501. Philos Trans R Soc Lond B Biol Sci. 1999. PMID: 10582243 Free PMC article. Review.
-
Transport of proteins to the yeast vacuole: autophagy, cytoplasm-to-vacuole targeting, and role of the vacuole in degradation.Semin Cell Dev Biol. 2000 Jun;11(3):173-9. doi: 10.1006/scdb.2000.0163. Semin Cell Dev Biol. 2000. PMID: 10906274 Review.
Cited by
-
An Atg10-like E2 enzyme is essential for cell cycle progression but not autophagy in Schizosaccharomyces pombe.Cell Cycle. 2013 Jan 15;12(2):271-7. doi: 10.4161/cc.23055. Epub 2012 Jan 15. Cell Cycle. 2013. PMID: 23255127 Free PMC article.
-
mTORC1 Activation Requires DRAM-1 by Facilitating Lysosomal Amino Acid Efflux.Mol Cell. 2019 Oct 3;76(1):163-176.e8. doi: 10.1016/j.molcel.2019.07.021. Epub 2019 Sep 3. Mol Cell. 2019. PMID: 31492633 Free PMC article.
-
Membrane recruitment of Aut7p in the autophagy and cytoplasm to vacuole targeting pathways requires Aut1p, Aut2p, and the autophagy conjugation complex.J Cell Biol. 2001 Jan 8;152(1):51-64. doi: 10.1083/jcb.152.1.51. J Cell Biol. 2001. PMID: 11149920 Free PMC article.
-
Marchantia polymorpha, a New Model Plant for Autophagy Studies.Front Plant Sci. 2019 Jul 17;10:935. doi: 10.3389/fpls.2019.00935. eCollection 2019. Front Plant Sci. 2019. PMID: 31379911 Free PMC article.
-
Functional Characterization of Ubiquitin-Like Core Autophagy Protein ATG12 in Dictyostelium discoideum.Cells. 2019 Jan 19;8(1):72. doi: 10.3390/cells8010072. Cells. 2019. PMID: 30669443 Free PMC article.
References
-
- Ausubel F, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Short Protocols in Molecular Biology. 3rd ed. New York: John Wiley & Sons; 1995.
-
- Baba M, Ohsumi M, Ohsumi Y. Analysis of the membrane structures involved in autophagy in yeast by freeze-replica method. Cell Struct Funct. 1995;20:465–471. - PubMed
-
- Boddy MN, Howe K, Etkin LD, Solomon E, Freemont PS. PIC1, a novel ubiquitin-like protein which interacts with the PML component of a multiprotein complex that is disrupted in acute promyelocytic leukemia. Oncogene. 1996;13:971–982. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

