CRM1-mediated recycling of snurportin 1 to the cytoplasm
- PMID: 10209022
- PMCID: PMC2133107
- DOI: 10.1083/jcb.145.2.255
CRM1-mediated recycling of snurportin 1 to the cytoplasm
Abstract
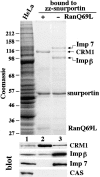
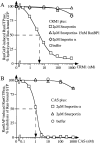
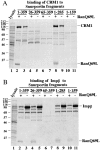
Importin beta is a major mediator of import into the cell nucleus. Importin beta binds cargo molecules either directly or via two types of adapter molecules, importin alpha, for import of proteins with a classical nuclear localization signal (NLS), or snurportin 1, for import of m3G-capped U snRNPs. Both adapters have an NH2-terminal importin beta-binding domain for binding to, and import by, importin beta, and both need to be returned to the cytoplasm after having delivered their cargoes to the nucleus. We have shown previously that CAS mediates export of importin alpha. Here we show that snurportin 1 is exported by CRM1, the receptor for leucine-rich nuclear export signals (NESs). However, the interaction of CRM1 with snurportin 1 differs from that with previously characterized NESs. First, CRM1 binds snurportin 1 50-fold stronger than the Rev protein and 5,000-fold stronger than the minimum Rev activation domain. Second, snurportin 1 interacts with CRM1 not through a short peptide but rather via a large domain that allows regulation of affinity. Strikingly, snurportin 1 has a low affinity for CRM1 when bound to its m3G-capped import substrate, and a high affinity when substrate-free. This mechanism appears crucial for productive import cycles as it can ensure that CRM1 only exports snurportin 1 that has already released its import substrate in the nucleus.
Figures



Similar articles
-
The importin β binding domain as a master regulator of nucleocytoplasmic transport.Biochim Biophys Acta. 2011 Sep;1813(9):1578-92. doi: 10.1016/j.bbamcr.2010.10.012. Epub 2010 Oct 26. Biochim Biophys Acta. 2011. PMID: 21029753 Free PMC article. Review.
-
Nucleocytoplasmic Shuttling of Porcine Parvovirus NS1 Protein Mediated by the CRM1 Nuclear Export Pathway and the Importin α/β Nuclear Import Pathway.J Virol. 2022 Jan 12;96(1):e0148121. doi: 10.1128/JVI.01481-21. Epub 2021 Oct 13. J Virol. 2022. PMID: 34643426 Free PMC article.
-
Interactions between HIV Rev and nuclear import and export factors: the Rev nuclear localisation signal mediates specific binding to human importin-beta.J Mol Biol. 1997 Dec 19;274(5):693-707. doi: 10.1006/jmbi.1997.1420. J Mol Biol. 1997. PMID: 9405152
-
Separate nuclear import pathways converge on the nucleoporin Nup153 and can be dissected with dominant-negative inhibitors.Curr Biol. 1998 Dec 17-31;8(25):1376-86. doi: 10.1016/s0960-9822(98)00018-9. Curr Biol. 1998. PMID: 9889100
-
Nucleocytoplasmic protein transport and recycling of Ran.Cell Struct Funct. 1999 Dec;24(6):425-33. doi: 10.1247/csf.24.425. Cell Struct Funct. 1999. PMID: 10698256 Review.
Cited by
-
p53 SUMOylation promotes its nuclear export by facilitating its release from the nuclear export receptor CRM1.Mol Biol Cell. 2013 Sep;24(17):2739-52. doi: 10.1091/mbc.E12-10-0771. Epub 2013 Jul 3. Mol Biol Cell. 2013. PMID: 23825024 Free PMC article.
-
Nuclear transport proteins: structure, function, and disease relevance.Signal Transduct Target Ther. 2023 Nov 10;8(1):425. doi: 10.1038/s41392-023-01649-4. Signal Transduct Target Ther. 2023. PMID: 37945593 Free PMC article. Review.
-
Novel-and Not So Novel-Inhibitors of the Multifunctional CRM1 Protein.Oncol Rev. 2024 Aug 5;18:1427497. doi: 10.3389/or.2024.1427497. eCollection 2024. Oncol Rev. 2024. PMID: 39161560 Free PMC article. Review.
-
The importin β binding domain as a master regulator of nucleocytoplasmic transport.Biochim Biophys Acta. 2011 Sep;1813(9):1578-92. doi: 10.1016/j.bbamcr.2010.10.012. Epub 2010 Oct 26. Biochim Biophys Acta. 2011. PMID: 21029753 Free PMC article. Review.
-
A supraphysiological nuclear export signal is required for parvovirus nuclear export.Mol Biol Cell. 2008 Jun;19(6):2544-52. doi: 10.1091/mbc.e08-01-0009. Epub 2008 Apr 2. Mol Biol Cell. 2008. PMID: 18385513 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

