Highly sensitive methods for quantitation of human immunodeficiency virus type 1 RNA from plasma, cells, and tissues
- PMID: 10203467
- PMCID: PMC84745
- DOI: 10.1128/JCM.37.5.1260-1264.1999
Highly sensitive methods for quantitation of human immunodeficiency virus type 1 RNA from plasma, cells, and tissues
Abstract
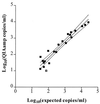
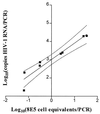
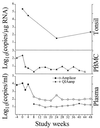
Precise and sensitive quantitation of viral RNA in specimens from human immunodeficiency virus (HIV) type 1 (HIV-1)-infected individuals has become an indispensable tool for the monitoring of the efficacy of highly active antiretroviral combination therapy. The present report describes reproducible and efficient protocols with enhanced sensitivity for quantitation of HIV-1 RNA from plasma, peripheral blood mononuclear cells, and tissues with Qiagen silica columns for RNA purification combined with the Roche Amplicor HIV-1 Monitor test for quantitative reverse transcription-PCR (RT-PCR). Extraction of RNA from 0.5 ml of plasma resulted in the detection of fewer than 20 HIV RNA copies/ml of plasma, equivalent to the centrifugation-based boosted RT-PCR assay. Silica extraction of cellular RNA resulted in the detection of fewer than 3 HIV-1 RNA copies/microg of total RNA. These techniques facilitate direct comparisons of viral loads between liquid and cellular specimens. Application of these sensitive methods may improve the assessment of the response to new antiretroviral regimens.
Figures
Similar articles
-
Clinical comparison of an enhanced-sensitivity branched-DNA assay and reverse transcription-PCR for quantitation of human immunodeficiency virus type 1 RNA in plasma.J Clin Microbiol. 1998 Mar;36(3):716-20. doi: 10.1128/JCM.36.3.716-720.1998. J Clin Microbiol. 1998. PMID: 9508301 Free PMC article.
-
Ultrasensitive reverse transcription-PCR assay for quantitation of human immunodeficiency virus type 1 RNA in plasma.J Clin Microbiol. 1998 Oct;36(10):2964-9. doi: 10.1128/JCM.36.10.2964-2969.1998. J Clin Microbiol. 1998. PMID: 9738051 Free PMC article.
-
Performance characteristics of the QUANTIPLEX HIV-1 RNA 3.0 assay for detection and quantitation of human immunodeficiency virus type 1 RNA in plasma.J Clin Microbiol. 2000 Aug;38(8):2837-45. doi: 10.1128/JCM.38.8.2837-2845.2000. J Clin Microbiol. 2000. PMID: 10921936 Free PMC article.
-
Clinical evaluation of an in-house reverse transcription-competitive PCR for quantitation of human immunodeficiency virus type 1 RNA in plasma.J Clin Microbiol. 1999 Feb;37(2):333-8. doi: 10.1128/JCM.37.2.333-338.1999. J Clin Microbiol. 1999. PMID: 9889213 Free PMC article.
-
Molecular-based methods for quantifying HIV viral load.AIDS Patient Care STDS. 2004 Feb;18(2):75-9. doi: 10.1089/108729104322802506. AIDS Patient Care STDS. 2004. PMID: 15006182 Review.
Cited by
-
Human immunodeficiency virus-specific CD8(+) T-cell responses do not predict viral growth and clearance rates during structured intermittent antiretroviral therapy.J Virol. 2002 Oct;76(20):10169-76. doi: 10.1128/jvi.76.20.10169-10176.2002. J Virol. 2002. PMID: 12239291 Free PMC article.
-
Cell-associated HIV-1 RNA predicts viral rebound and disease progression after discontinuation of temporary early ART.JCI Insight. 2020 Mar 26;5(6):e134196. doi: 10.1172/jci.insight.134196. JCI Insight. 2020. PMID: 32097124 Free PMC article. Clinical Trial.
-
Biphasic decay kinetics suggest progressive slowing in turnover of latently HIV-1 infected cells during antiretroviral therapy.Retrovirology. 2008 Nov 26;5:107. doi: 10.1186/1742-4690-5-107. Retrovirology. 2008. PMID: 19036147 Free PMC article.
-
Barriers for HIV Cure: The Latent Reservoir.AIDS Res Hum Retroviruses. 2018 Sep;34(9):739-759. doi: 10.1089/AID.2018.0118. Epub 2018 Aug 28. AIDS Res Hum Retroviruses. 2018. PMID: 30056745 Free PMC article. Review.
-
Entirely automated quantification of human immunodeficiency virus type 1 (HIV-1) RNA in plasma by using the ultrasensitive COBAS AMPLICOR HIV-1 monitor test and RNA purification on the MagNA pure LC instrument.J Clin Microbiol. 2003 Mar;41(3):1248-51. doi: 10.1128/JCM.41.3.1248-1251.2003. J Clin Microbiol. 2003. PMID: 12624059 Free PMC article.
References
-
- Ausubel S M, Brent R M, Kingston R E, Moore D D, Seldmann J G, Smith J G, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1987.
-
- Cavert W, Notermans D W, Staskus K, Wietgrefe S W, Zupancic M, Gebhard K, Henr K, Zhang Z Q, Mills R, McDade H, Goudsmit J, Danner S A, Haase A T. Kinetics of response in lymphoid tissues to antiretroviral therapy of HIV-1 infection. Science. 1997;276:960–964. - PubMed
-
- Finzi D, Hermankova M, Pierson T, Carruth L M, Buck C, Chaisson R E, Quinn T C, Chadwick K, Margolick J, Brookmeyer R, Gallant J, Markowitz M, Ho D D, Richman D D, Siliciano R F. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science. 1997;278:1295–1300. - PubMed
-
- Gulick R M, Mellors J W, Havlir D, Eron J J, Gonzalez C, McMahon D, Richman D D, Valentine F T, Jonas L, Meibohm A, Emini E A, Chodakewitz J A. Treatment with indinavir, zidovudine, and lamivudine in adults with human immunodeficiency virus infection and prior antiretroviral therapy. N Engl J Med. 1997;337:734–739. - PubMed
-
- Hahn B H, Shaw G M, Arya S K, Popovic M, Gallo R C, Wong Staal F. Molecular cloning and characterization of the HTLV-III virus associated with AIDS. Nature. 1984;312:166–169. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

