Human RNA-specific adenosine deaminase ADAR1 transcripts possess alternative exon 1 structures that initiate from different promoters, one constitutively active and the other interferon inducible
- PMID: 10200312
- PMCID: PMC16382
- DOI: 10.1073/pnas.96.8.4621
Human RNA-specific adenosine deaminase ADAR1 transcripts possess alternative exon 1 structures that initiate from different promoters, one constitutively active and the other interferon inducible
Abstract
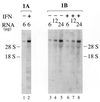
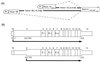
RNA-specific adenosine deaminase (ADAR1) catalyzes the deamination of adenosine to inosine in viral and cellular RNAs. Two size forms of the ADAR1 editing enzyme are known, an IFN-inducible approximately 150-kDa protein and a constitutively expressed N-terminally truncated approximately 110-kDa protein. We have now identified alternative exon 1 structures of human ADAR1 transcripts that initiate from unique promoters, one constitutively expressed and the other IFN inducible. Cloning and sequence analyses of 5'-rapid amplification of cDNA ends (RACE) cDNAs from human placenta established a linkage between exon 2 of ADAR1 and two alternative exon 1 structures, designated herein as exon 1A and exon 1B. Analysis of RNA isolated from untreated and IFN-treated human amnion cells demonstrated that exon 1B-exon 2 transcripts were synthesized in the absence of IFN and were not significantly altered in amount by IFN treatment. By contrast, exon 1A-exon 2 transcripts were IFN inducible. Transient transfection analysis with reporter constructs led to the identification of two functional promoters, designated PC and PI. Exon 1B transcripts were initiated from the PC promoter whose activity in transient transfection reporter assays was not increased by IFN treatment. The 107-nt exon 1B mapped 14.5 kb upstream of exon 2. The 201-nt exon 1A that mapped 5.4 kb upstream of exon 2 was initiated from the interferon-inducible PI promoter. These results suggest that two promoters, one IFN inducible and the other not, initiate transcription of the ADAR1 gene, and that alternative splicing of unique exon 1 structures to a common exon 2 junction generates RNA transcripts with the deduced coding capacity for either the constitutively expressed approximately 110-kDa ADAR1 protein (exon 1B) or the interferon-induced approximately 150-kDa ADAR1 protein (exon 1A).
Figures





Similar articles
-
Expression of interferon-inducible RNA adenosine deaminase ADAR1 during pathogen infection and mouse embryo development involves tissue-selective promoter utilization and alternative splicing.J Biol Chem. 2005 Apr 15;280(15):15020-8. doi: 10.1074/jbc.M500476200. Epub 2005 Jan 25. J Biol Chem. 2005. PMID: 15677478
-
Human RNA-specific adenosine deaminase (ADAR1) gene specifies transcripts that initiate from a constitutively active alternative promoter.Gene. 2000 Nov 27;258(1-2):165-72. doi: 10.1016/s0378-1119(00)00368-1. Gene. 2000. PMID: 11111054
-
Characterization of the 5'-flanking region of the human RNA-specific adenosine deaminase ADAR1 gene and identification of an interferon-inducible ADAR1 promoter.Gene. 1999 Mar 18;229(1-2):203-13. doi: 10.1016/s0378-1119(99)00017-7. Gene. 1999. PMID: 10095120
-
An RNA editor, adenosine deaminase acting on double-stranded RNA (ADAR1).J Interferon Cytokine Res. 2014 Jun;34(6):437-46. doi: 10.1089/jir.2014.0001. J Interferon Cytokine Res. 2014. PMID: 24905200 Free PMC article. Review.
-
Adenosine deaminases acting on RNA, RNA editing, and interferon action.J Interferon Cytokine Res. 2011 Jan;31(1):99-117. doi: 10.1089/jir.2010.0097. Epub 2010 Dec 23. J Interferon Cytokine Res. 2011. PMID: 21182352 Free PMC article. Review.
Cited by
-
Regulatory factors governing adenosine-to-inosine (A-to-I) RNA editing.Biosci Rep. 2015 Mar 31;35(2):e00182. doi: 10.1042/BSR20140190. Biosci Rep. 2015. PMID: 25662729 Free PMC article. Review.
-
Endogenous Double-Stranded RNA.Noncoding RNA. 2021 Feb 19;7(1):15. doi: 10.3390/ncrna7010015. Noncoding RNA. 2021. PMID: 33669629 Free PMC article. Review.
-
MxA transcripts with distinct first exons and modulation of gene expression levels by single-nucleotide polymorphisms in human bronchial epithelial cells.Immunogenetics. 2013 Feb;65(2):107-14. doi: 10.1007/s00251-012-0663-8. Epub 2012 Nov 18. Immunogenetics. 2013. PMID: 23160781 Free PMC article.
-
Hepatitis D Virus Infection of Mice Expressing Human Sodium Taurocholate Co-transporting Polypeptide.PLoS Pathog. 2015 Apr 22;11(4):e1004840. doi: 10.1371/journal.ppat.1004840. eCollection 2015 Apr. PLoS Pathog. 2015. PMID: 25902143 Free PMC article.
-
Effects of length and location on the cellular response to double-stranded RNA.Microbiol Mol Biol Rev. 2004 Sep;68(3):432-52, table of contents. doi: 10.1128/MMBR.68.3.432-452.2004. Microbiol Mol Biol Rev. 2004. PMID: 15353564 Free PMC article. Review.
References
-
- Simpson L, Emeson R B. Annu Rev Neurosci. 1996;19:27–52. - PubMed
-
- Rueter S M, Emeson R B. Modifications and Editing of RNA. Washington, D.C.: Am. Soc. Microbiol.; 1998. pp. 343–361.
-
- Seeburg P H, Higuchi M, Sprengel R. Brain Res Rev. 1998;26:217–229. - PubMed
-
- Bass B L. Trends Biochem Sci. 1997;22:157–162. - PubMed
-
- Burns C M, Chu H, Rueter S M, Hutchinson L K, Canton H, Sanders-Bush E, Emeson R B. Nature (London) 1997;387:303–308. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

