Cyanovirin-N binds to gp120 to interfere with CD4-dependent human immunodeficiency virus type 1 virion binding, fusion, and infectivity but does not affect the CD4 binding site on gp120 or soluble CD4-induced conformational changes in gp120
- PMID: 10196334
- PMCID: PMC104217
- DOI: 10.1128/JVI.73.5.4360-4371.1999
Cyanovirin-N binds to gp120 to interfere with CD4-dependent human immunodeficiency virus type 1 virion binding, fusion, and infectivity but does not affect the CD4 binding site on gp120 or soluble CD4-induced conformational changes in gp120
Abstract
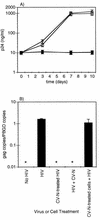
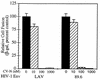
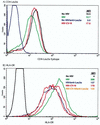
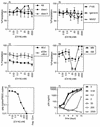
Cyanovirin-N (CV-N), an 11-kDa protein isolated from the cyanobacterium Nostoc ellipsosporum, potently inactivates diverse strains of human immunodeficiency virus type 1 (HIV-1), HIV-2, and simian immunodeficiency virus. While it has been well established that the viral surface envelope glycoprotein gp120 is a molecular target of CV-N, the detailed mechanism of action is of further interest. We compared matched native and CV-N-treated virus preparations in a panel of assays that measure viral replication, assessing successive stages of the viral life cycle. CV-N-treated virions failed to infect cells as detected by p24 production and quantitative PCR for HIV-1 reverse transcription products, whereas treatment of the target cells did not block infection, confirming that CV-N acts at the level of the virus, not the target cell, to abort the initial infection process. Compared to native HIV-1 preparations, CV-N-treated HIV-1 virions showed impaired CD4-dependent binding to CD4(+) T cells and did not mediate "fusion from without" of CD4(+) target cells. CV-N also blocked HIV envelope glycoprotein Env-induced, CD4-dependent cell-cell fusion. Mapping studies with monoclonal antibodies (MAbs) to defined epitopes on the HIV-1 envelope glycoprotein indicated that CV-N binds to gp120 in a manner that does not occlude or alter the CD4 binding site or V3 loop or other domains on gp120 recognized by defined MAbs and does not interfere with soluble CD4-induced conformational changes in gp120. Binding of CV-N to soluble gp120 or virions inhibited subsequent binding of the unique neutralizing MAb 2G12, which recognizes a glycosylation-dependent epitope. However, prior binding of 2G12 MAb to gp120 did not block subsequent binding by CV-N. These results help clarify the mechanism of action of CV-N and suggest that the compound may act in part by preventing essential interactions between the envelope glycoprotein and target cell receptors. This proposed mechanism is consistent with the extensive activity profile of CV-N against numerous isolates of HIV-1 and other lentiviruses and supports the potential broad utility of this protein as a microbicide to prevent the sexual transmission of HIV.
Figures




Similar articles
-
Cyanovirin-N, a potent human immunodeficiency virus-inactivating protein, blocks both CD4-dependent and CD4-independent binding of soluble gp120 (sgp120) to target cells, inhibits sCD4-induced binding of sgp120 to cell-associated CXCR4, and dissociates bound sgp120 from target cells.Antimicrob Agents Chemother. 2001 Mar;45(3):664-72. doi: 10.1128/AAC.45.3.664-672.2001. Antimicrob Agents Chemother. 2001. PMID: 11181340 Free PMC article.
-
Multiple antiviral activities of cyanovirin-N: blocking of human immunodeficiency virus type 1 gp120 interaction with CD4 and coreceptor and inhibition of diverse enveloped viruses.J Virol. 2000 May;74(10):4562-9. doi: 10.1128/jvi.74.10.4562-4569.2000. J Virol. 2000. PMID: 10775592 Free PMC article.
-
Conformational changes affecting the V3 and CD4-binding domains of human immunodeficiency virus type 1 gp120 associated with env processing and with binding of ligands to these sites.J Virol. 1993 Sep;67(9):5692-7. doi: 10.1128/JVI.67.9.5692-5697.1993. J Virol. 1993. PMID: 7688827 Free PMC article.
-
Properties of cyanovirin-N (CV-N): inactivation of HIV-1 by sessile cyanovirin-N (sCV-N).Dev Biol (Basel). 2000;102:141-8. Dev Biol (Basel). 2000. PMID: 10794101 Review.
-
CD4 activation of HIV fusion.Int J Cell Cloning. 1992 Nov;10(6):323-32. doi: 10.1002/stem.5530100603. Int J Cell Cloning. 1992. PMID: 1281202 Review.
Cited by
-
The conserved residue Arg46 in the N-terminal heptad repeat domain of HIV-1 gp41 is critical for viral fusion and entry.PLoS One. 2012;7(9):e44874. doi: 10.1371/journal.pone.0044874. Epub 2012 Sep 7. PLoS One. 2012. PMID: 22970321 Free PMC article.
-
Anti-HIV activity of defective cyanovirin-N mutants is restored by dimerization.J Biol Chem. 2010 Apr 23;285(17):13057-65. doi: 10.1074/jbc.M109.094938. Epub 2010 Feb 10. J Biol Chem. 2010. PMID: 20147291 Free PMC article.
-
Cyanovirin-N, a potent human immunodeficiency virus-inactivating protein, blocks both CD4-dependent and CD4-independent binding of soluble gp120 (sgp120) to target cells, inhibits sCD4-induced binding of sgp120 to cell-associated CXCR4, and dissociates bound sgp120 from target cells.Antimicrob Agents Chemother. 2001 Mar;45(3):664-72. doi: 10.1128/AAC.45.3.664-672.2001. Antimicrob Agents Chemother. 2001. PMID: 11181340 Free PMC article.
-
Surfactant Protein D modulates HIV infection of both T-cells and dendritic cells.PLoS One. 2013;8(3):e59047. doi: 10.1371/journal.pone.0059047. Epub 2013 Mar 18. PLoS One. 2013. PMID: 23527085 Free PMC article.
-
Development of an HIV-1 Microbicide Based on Caulobacter crescentus: Blocking Infection by High-Density Display of Virus Entry Inhibitors.PLoS One. 2013 Jun 19;8(6):e65965. doi: 10.1371/journal.pone.0065965. Print 2013. PLoS One. 2013. PMID: 23840383 Free PMC article.
References
-
- Alkhatib, G., and E. A. Berger. Unpublished data.
-
- Arthos J, Deen K C, Chaikin M A, Fornwald J A, Sathe G, Sattentau Q J, Clapham P R, Weiss R A, McDougal J S, Pietropaolo C, et al. Identification of the residues in human CD4 critical for the binding of HIV. Cell. 1989;57:469–481. - PubMed
-
- Arthur L O, Bess J W, Jr, Sowder R C D, Benveniste R E, Mann D L, Chermann J C, Henderson L E. Cellular proteins bound to immunodeficiency viruses: implications for pathogenesis and vaccines. Science. 1992;258:1935–1938. - PubMed
-
- Attanasio R, Diley D, Buck D, Maino V C, Lohman K L, Kanda P, Kennedy R C. Structural characterization of a cross-reactive idiotype shared by monoclonal antibodies specific for the human CD4 molecule. J Biol Chem. 1991;266:14611–14619. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

