High-level variability in the ORF-K1 membrane protein gene at the left end of the Kaposi's sarcoma-associated herpesvirus genome defines four major virus subtypes and multiple variants or clades in different human populations
- PMID: 10196312
- PMCID: PMC104195
- DOI: 10.1128/JVI.73.5.4156-4170.1999
High-level variability in the ORF-K1 membrane protein gene at the left end of the Kaposi's sarcoma-associated herpesvirus genome defines four major virus subtypes and multiple variants or clades in different human populations
Abstract
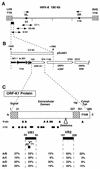
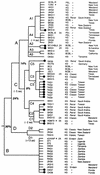
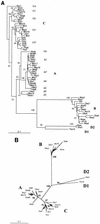
Infection with Kaposi's sarcoma (KS)-associated herpesvirus (KSHV) or human herpesvirus 8 (HHV8) is common in certain parts of Africa, the Middle East, and the Mediterranean, but is rare elsewhere, except in AIDS patients. Nevertheless, HHV8 DNA is found consistently in nearly all classical, endemic, transplant and AIDS-associated KS lesions as well as in some rare AIDS-associated lymphomas. The concept that HHV8 genomes fall into several distinct subgroups has been confirmed and refined by PCR DNA sequence analysis of the ORF-K1 gene encoding a highly variable glycoprotein related to the immunoglobulin receptor family that maps at the extreme left-hand end of the HHV-8 genome. Among more than 60 different tumor samples from the United States, central Africa, Saudi Arabia, Taiwan, and New Zealand, amino acid substitutions were found at a total of 62% of the 289 amino acid positions. These variations defined four major subtypes and 13 distinct variants or clades similar to those found for the HIV ENV protein. The B and D subtype ORF-K1 proteins differ from the A and C subtypes by 30 and 24%, respectively, whereas A and C differ from each other by 15%. In all cases tested, multiple samples from the same patient were identical. Examples of the B subtype were found almost exclusively in KS patients from Africa or of African heritage, whereas the rare D subtypes were found only in KS patients of Pacific Island heritage. In contrast, C subtypes were found predominantly in classic KS and in iatrogenic and AIDS KS in the Middle East and Asia, whereas U.S. AIDS KS samples were primarily A1, A4, and C3 variants. We conclude that this unusually high diversity, in which 85% of the nucleotide changes lead to amino acid changes, reflects some unknown powerful biological selection process that has been acting preferentially on this early lytic cycle membrane signalling protein. Two distinct levels of ORF-K1 variability are recognizable. Subtype-specific variability indicative of long-term evolutionary divergence is both spread throughout the protein as well as concentrated within two 40-amino-acid extracellular domain variable regions (VR1 and VR2), whereas intratypic variability localizes predominantly within a single 25-amino-acid hypervariable Cys bridge loop and apparently represents much more recent changes that have occurred even within specific clades. In contrast, numerous extracellular domain glycosylation sites and Cys bridge residues as well as the ITAM motif in the cytoplasmic domain are fully conserved. Overall, we suggest that rather than being a newly acquired human pathogen, HHV8 is an ancient human virus that is preferentially transmitted in a familial fashion and is difficult to transmit horizontally in the absence of immunosuppression. The division into the four major HHV8 subgroups is probably the result of isolation and founder effects associated with the history of migration of modern human populations out of Africa over the past 35,000 to 60,000 years.
Figures







Similar articles
-
Genotypic analysis at multiple loci across Kaposi's sarcoma herpesvirus (KSHV) DNA molecules: clustering patterns, novel variants and chimerism.J Clin Virol. 2002 Jan;23(3):119-48. doi: 10.1016/s1386-6532(01)00205-0. J Clin Virol. 2002. PMID: 11595592
-
Comparison of genetic variability at multiple loci across the genomes of the major subtypes of Kaposi's sarcoma-associated herpesvirus reveals evidence for recombination and for two distinct types of open reading frame K15 alleles at the right-hand end.J Virol. 1999 Aug;73(8):6646-60. doi: 10.1128/JVI.73.8.6646-6660.1999. J Virol. 1999. PMID: 10400762 Free PMC article.
-
Human herpesvirus type 8 genotypes in iatrogenic, classic and AIDS-associated Kaposi's sarcoma from Greece.Anticancer Res. 2004 May-Jun;24(3a):1597-602. Anticancer Res. 2004. PMID: 15274328
-
KSHV strains: the origins and global spread of the virus.Semin Cancer Biol. 1999 Jun;9(3):187-99. doi: 10.1006/scbi.1998.0116. Semin Cancer Biol. 1999. PMID: 10343070 Review.
-
Epidemiology and clinical characteristics of classic Kaposi's sarcoma, seroprevalence, and variants of human herpesvirus 8 in South America: a critical review of an old disease.Int J Infect Dis. 2005 Sep;9(5):239-50. doi: 10.1016/j.ijid.2005.02.004. Int J Infect Dis. 2005. PMID: 16095940 Review.
Cited by
-
Two distinct gamma-2 herpesviruses in African green monkeys: a second gamma-2 herpesvirus lineage among old world primates?J Virol. 2000 Feb;74(3):1572-7. doi: 10.1128/jvi.74.3.1572-1577.2000. J Virol. 2000. PMID: 10627572 Free PMC article.
-
Phylogenetic analysis of Ostreococcus virus sequences from the Patagonian Coast.Virus Genes. 2012 Oct;45(2):316-26. doi: 10.1007/s11262-012-0762-5. Epub 2012 Jun 7. Virus Genes. 2012. PMID: 22674355
-
Model-based inference of recombination hotspots in a highly variable oncogene [corrected].J Mol Evol. 2004 Mar;58(3):239-51. doi: 10.1007/s00239-003-2543-1. J Mol Evol. 2004. PMID: 15045480
-
Molecular analysis of human herpesvirus 8 by using single nucleotide polymorphisms in open reading frame 26.J Clin Microbiol. 2003 Jun;41(6):2492-7. doi: 10.1128/JCM.41.6.2492-2497.2003. J Clin Microbiol. 2003. PMID: 12791871 Free PMC article.
-
Molecular evolution of the gamma-Herpesvirinae.Philos Trans R Soc Lond B Biol Sci. 2001 Apr 29;356(1408):421-35. doi: 10.1098/rstb.2000.0775. Philos Trans R Soc Lond B Biol Sci. 2001. PMID: 11313003 Free PMC article. Review.
References
-
- Alagouzoulou, L., J.-C. Zong, and G. S. Hayward. Unpublished data.
-
- Ambinder R F, Newman C, Hayward G S, Biggar R, Melbye M, Kestens L, Van Marck E, Piot P, Gigase P, Wright P B, Quinn T C. Lack of association of cytomegalovirus with endemic African Kaposi’s sarcoma. J Infect Dis. 1987;156:193–197. - PubMed
-
- Arvanitakis L, Mesri E A, Nador R G, Said J W, Asch A S, Knowles D M, Cesarman E. Establishment and characterization of a primary effusion (body cavity-based) lymphoma cell line (BC-3) harboring Kaposi’s sarcoma associated herpesvirus (KSHV/HHV-8) in the absence of Epstein-Barr virus. Blood. 1996;88:2648–2654. - PubMed
-
- Beaufils P, Choquet D, Mamoun R Z, Malissen B. The (YXXL/I)2 signalling motif found in the cytoplasmic segments of the bovine leukaemia virus envelope protein and Epstein-Barr virus latent membrane protein 2A can elicit early and late lymphocyte activation events. EMBO J. 1993;12:5105–5112. - PMC - PubMed
-
- Beral V. Epidemiology of Kaposi’s sarcoma. Cancer Surv. 1991;10:5–22. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

