Esa1p is an essential histone acetyltransferase required for cell cycle progression
- PMID: 10082517
- PMCID: PMC84044
- DOI: 10.1128/MCB.19.4.2515
Esa1p is an essential histone acetyltransferase required for cell cycle progression
Abstract
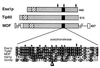
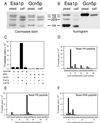
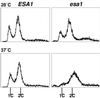
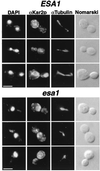
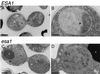
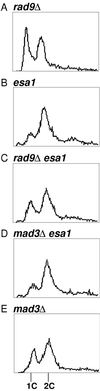
Histones are dynamically modified during chromatin assembly, as specific transcriptional patterns are established, and during mitosis and development. Modifications include acetylation, phosphorylation, ubiquitination, methylation, and ADP-ribosylation, but the biological significance of each of these is not well understood. For example, distinct acetylation patterns correlate with nucleosome formation and with transcriptionally activated or silenced chromatin, yet mutations in genes encoding several yeast histone acetyltransferase (HAT) activities result in either no cellular phenotype or only modest growth defects. Here we report characterization of ESA1, an essential gene that is a member of the MYST family that includes two yeast silencing genes, human genes associated with leukemia and with the human immunodeficiency virus type 1 Tat protein, and Drosophila mof, a gene essential for male dosage compensation. Esa1p acetylates histones in a pattern distinct from those of other yeast enzymes, and temperature-sensitive mutant alleles abolish enzymatic activity in vitro and result in partial loss of an acetylated isoform of histone H4 in vivo. Strains carrying these mutations are also blocked in the cell cycle such that at restrictive temperatures, esa1 mutants succeed in replicating their DNA but fail to proceed normally through mitosis and cytokinesis. Recent studies show that Esa1p enhances transcription in vitro and thus may modulate expression of genes important for cell cycle control. These observations therefore link an essential HAT activity to cell cycle progression, potentially through discrete transcriptional regulatory events.
Figures

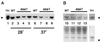

Similar articles
-
NuA4, an essential transcription adaptor/histone H4 acetyltransferase complex containing Esa1p and the ATM-related cofactor Tra1p.EMBO J. 1999 Sep 15;18(18):5108-19. doi: 10.1093/emboj/18.18.5108. EMBO J. 1999. PMID: 10487762 Free PMC article.
-
Histone H3 specific acetyltransferases are essential for cell cycle progression.Genes Dev. 2001 Dec 1;15(23):3144-54. doi: 10.1101/gad.931401. Genes Dev. 2001. PMID: 11731478 Free PMC article.
-
ESA1 is a histone acetyltransferase that is essential for growth in yeast.Proc Natl Acad Sci U S A. 1998 Mar 31;95(7):3561-5. doi: 10.1073/pnas.95.7.3561. Proc Natl Acad Sci U S A. 1998. PMID: 9520405 Free PMC article.
-
Nuclear matrix, dynamic histone acetylation and transcriptionally active chromatin.Mol Biol Rep. 1997 Aug;24(3):197-207. doi: 10.1023/a:1006811817247. Mol Biol Rep. 1997. PMID: 9291093 Review.
-
The diverse functions of histone acetyltransferase complexes.Trends Genet. 2003 Jun;19(6):321-9. doi: 10.1016/S0168-9525(03)00115-X. Trends Genet. 2003. PMID: 12801725 Review.
Cited by
-
Evidence for monomeric actin function in INO80 chromatin remodeling.Nat Struct Mol Biol. 2013 Apr;20(4):426-32. doi: 10.1038/nsmb.2529. Epub 2013 Mar 24. Nat Struct Mol Biol. 2013. PMID: 23524535 Free PMC article.
-
Drosophila dCBP is involved in establishing the DNA replication checkpoint.Mol Cell Biol. 2007 Jan;27(1):135-46. doi: 10.1128/MCB.01283-06. Epub 2006 Oct 16. Mol Cell Biol. 2007. PMID: 17043110 Free PMC article.
-
Schizosaccharomyces pombe mst2+ encodes a MYST family histone acetyltransferase that negatively regulates telomere silencing.Mol Cell Biol. 2005 Oct;25(20):8887-903. doi: 10.1128/MCB.25.20.8887-8903.2005. Mol Cell Biol. 2005. PMID: 16199868 Free PMC article.
-
The Saccharomyces cerevisiae Piccolo NuA4 histone acetyltransferase complex requires the Enhancer of Polycomb A domain and chromodomain to acetylate nucleosomes.Mol Cell Biol. 2005 Jul;25(13):5535-42. doi: 10.1128/MCB.25.13.5535-5542.2005. Mol Cell Biol. 2005. PMID: 15964809 Free PMC article.
-
Histone acetylation at promoters is differentially affected by specific activators and repressors.Mol Cell Biol. 2001 Apr;21(8):2726-35. doi: 10.1128/MCB.21.8.2726-2735.2001. Mol Cell Biol. 2001. PMID: 11283252 Free PMC article.
References
-
- Allen J B, Zhou Z, Siede W, Freiberg E C, Elledge S J. The SAD1/RAD53 protein kinase controls multiple checkpoints and DNA damage-induced transcription in yeast. Genes Dev. 1994;8:652–665. - PubMed
-
- Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Bannister A J, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
