Intermediates in formation and activity of the RNA polymerase II preinitiation complex: holoenzyme recruitment and a postrecruitment role for the TATA box and TFIIB
- PMID: 9887099
- PMCID: PMC316368
- DOI: 10.1101/gad.13.1.49
Intermediates in formation and activity of the RNA polymerase II preinitiation complex: holoenzyme recruitment and a postrecruitment role for the TATA box and TFIIB
Abstract
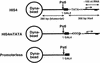
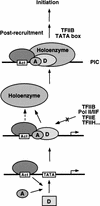
Assembly and activity of yeast RNA polymerase II (Pol II) preinitiation complexes (PIC) was investigated with an immobilized promoter assay and extracts made from wild-type cells and from cells containing conditional mutations in components of the Pol II machinery. We describe the following findings: (1) In one step, TFIID and TFIIA assemble at the promoter independently of holoenzyme. In another step, holoenzyme is recruited to the promoter. Mutations in the CTD of Pol II, Srb2, Srb4, and Srb5, and two mutations in TFIIB disrupt recruitment of all holoenzyme components tested without affecting TFIID and TFIIA recruitment. These results indicate that the stepwise assembly pathway is blocked after TFIID/TFIIA binding. (2) Both the Gal4-AH and Gal4-VP16 activators stimulate formation of active PICs by increasing the extent of PIC formation. The Gal4-AH activator stimulated PIC formation by enhancing the binding of TFIID and TFIIA, whereas Gal4-VP16 could enhance the recruitment of TFIID, TFIIA, and holoenzyme. (3) Extracts deficient in TFIIA activity showed reduced assembly of all PIC components. These and other results suggest that TFIIA acts at an early step by enhancing the stable recruitment of TFIID. (4) An extract containing the TFIIB mutant E62G, had no defect in PIC formation, but had a severe defect in transcription. Similarly, mutation of the TATA box reduced PIC formation only two- to fourfold, but severely compromised transcription. These results demonstate an involvement of TFIIB and the TATA box in one or more steps after recruitment of factors to the promoter.
Figures

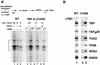
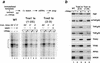

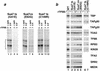
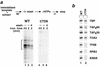

Similar articles
-
Virtually unidirectional binding of TBP to the AdMLP TATA box within the quaternary complex with TFIIA and TFIIB.Chem Biol. 2000 Aug;7(8):601-10. doi: 10.1016/s1074-5521(00)00009-0. Chem Biol. 2000. PMID: 11048951
-
Recruitment of TBP or TFIIB to a promoter proximal position leads to stimulation of RNA polymerase II transcription without activator proteins both in vivo and in vitro.Biochem Biophys Res Commun. 1999 Mar 5;256(1):45-51. doi: 10.1006/bbrc.1999.0280. Biochem Biophys Res Commun. 1999. PMID: 10066420
-
Structural insights into assembly of transcription preinitiation complex.Curr Opin Struct Biol. 2022 Aug;75:102404. doi: 10.1016/j.sbi.2022.102404. Epub 2022 Jun 11. Curr Opin Struct Biol. 2022. PMID: 35700575 Review.
-
RNA polymerase II components and Rrn7 form a preinitiation complex on the HomolD box to promote ribosomal protein gene expression in Schizosaccharomyces pombe.FEBS J. 2017 Feb;284(4):615-633. doi: 10.1111/febs.14006. Epub 2017 Feb 5. FEBS J. 2017. PMID: 28060464
-
Mechanisms of transcriptional activation in vivo: two steps forward.Trends Genet. 1996 Aug;12(8):311-5. doi: 10.1016/0168-9525(96)10028-7. Trends Genet. 1996. PMID: 8783941 Review.
Cited by
-
The transcription elongation factor TFIIS is a component of RNA polymerase II preinitiation complexes.Proc Natl Acad Sci U S A. 2007 Oct 9;104(41):16068-73. doi: 10.1073/pnas.0704573104. Epub 2007 Oct 3. Proc Natl Acad Sci U S A. 2007. PMID: 17913884 Free PMC article.
-
The linker domain of basal transcription factor TFIIB controls distinct recruitment and transcription stimulation functions.Nucleic Acids Res. 2011 Jan;39(2):464-74. doi: 10.1093/nar/gkq809. Epub 2010 Sep 17. Nucleic Acids Res. 2011. PMID: 20851833 Free PMC article.
-
TFIIA interacts with TFIID via association with TATA-binding protein and TAF40.Mol Cell Biol. 2001 Mar;21(5):1737-46. doi: 10.1128/MCB.21.5.1737-1746.2001. Mol Cell Biol. 2001. PMID: 11238911 Free PMC article.
-
Interdependent interactions between TFIIB, TATA binding protein, and DNA.Mol Cell Biol. 2002 Dec;22(24):8735-43. doi: 10.1128/MCB.22.24.8735-8743.2002. Mol Cell Biol. 2002. PMID: 12446790 Free PMC article.
-
A triad of subunits from the Gal11/tail domain of Srb mediator is an in vivo target of transcriptional activator Gcn4p.Mol Cell Biol. 2004 Aug;24(15):6871-86. doi: 10.1128/MCB.24.15.6871-6886.2004. Mol Cell Biol. 2004. PMID: 15254252 Free PMC article.
References
-
- Auble DT, Hahn S. An ATP-dependent inhibitor of TBP binding to DNA. Genes & Dev. 1993;7:844–856. - PubMed
-
- Bryant GO, Martel LS, Burley SK, Berk AJ. Radical mutations reveal TATA-box binding protein surfaces required for activated transcription in vivo. Genes & Dev. 1996;10:2491–2504. - PubMed
-
- Bushnell DA, Bamdad C, Kornberg RD. A minimal set of RNA polymerase II transcription protein interactions. J Biol Chem. 1996;271:20170–20174. - PubMed
-
- Chi T, Carey M. Assembly of the isomerized TFIIA—TFIID—TATA ternary complex is necessary and sufficient for gene activation. Genes & Dev. 1996;10:2540–2550. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
