Proapoptotic activity of Caenorhabditis elegans CED-4 protein in Drosophila: implicated mechanisms for caspase activation
- PMID: 9874786
- PMCID: PMC15107
- DOI: 10.1073/pnas.96.1.145
Proapoptotic activity of Caenorhabditis elegans CED-4 protein in Drosophila: implicated mechanisms for caspase activation
Abstract
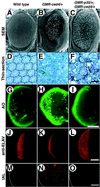
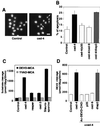
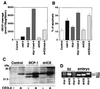
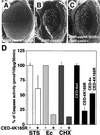
CED-4 protein plays an important role in the induction of programmed cell death in Caenorhabditis elegans through the activation of caspases. However, the precise mechanisms by which it activates caspases remain unknown. To investigate the conservation of CED-4 function in evolution, transgenic Drosophila lines that express CED-4 in the compound eye were generated. Ectopic expression of CED-4 in the eyes induced massive apoptotic cell death through caspase activation. An ATP-binding site (P-loop) mutation in CED-4 (K165R) causes a loss of function in its ability to activate Drosophila caspase, and an ATPase inhibitor blocks the CED-4-dependent caspase activity in Drosophila S2 cells. Immunoprecipitation analysis showed that both CED-4 and CED-4 (K165R) bind directly to Drosophila caspase drICE, and the overexpression of CED-4 (K165R) inhibits CED-4-, ecdysone-, or cycloheximide-dependent caspase activation in S2 cells. Furthermore, CED-4 (K165R) partially prevented cell death induced by CED-4 in Drosophila compound eyes. Thus, CED-4 function is evolutionarily conserved in Drosophila, and the molecular mechanisms by which CED-4 activates caspases might require ATP binding and direct interaction with the caspases.
Figures

Similar articles
-
Translocation of C. elegans CED-4 to nuclear membranes during programmed cell death.Science. 2000 Feb 25;287(5457):1485-9. doi: 10.1126/science.287.5457.1485. Science. 2000. PMID: 10688797
-
Interaction of CED-4 with CED-3 and CED-9: a molecular framework for cell death.Science. 1997 Feb 21;275(5303):1122-6. doi: 10.1126/science.275.5303.1122. Science. 1997. PMID: 9027312
-
Drob-1, a Drosophila member of the Bcl-2/CED-9 family that promotes cell death.Proc Natl Acad Sci U S A. 2000 Jan 18;97(2):662-7. doi: 10.1073/pnas.97.2.662. Proc Natl Acad Sci U S A. 2000. PMID: 10639136 Free PMC article.
-
Ced-3/ICE: evolutionarily conserved regulation of cell death.Bioessays. 1994 Jun;16(6):387-9. doi: 10.1002/bies.950160604. Bioessays. 1994. PMID: 8080427 Review. No abstract available.
-
2:1 Stoichiometry of the CED-4-CED-9 complex and the tetrameric CED-4: insights into the regulation of CED-3 activation.Cell Cycle. 2006 Jan;5(1):31-4. doi: 10.4161/cc.5.1.2263. Epub 2006 Jan 18. Cell Cycle. 2006. PMID: 16294007 Review.
Cited by
-
Caspase-9 activation revealed by semaphorin 7A cleavage is independent of apoptosis in the aged olfactory bulb.J Neurosci. 2009 Sep 9;29(36):11385-92. doi: 10.1523/JNEUROSCI.4780-08.2009. J Neurosci. 2009. PMID: 19741144 Free PMC article.
-
Eiger, a TNF superfamily ligand that triggers the Drosophila JNK pathway.EMBO J. 2002 Jun 17;21(12):3009-18. doi: 10.1093/emboj/cdf306. EMBO J. 2002. PMID: 12065414 Free PMC article.
-
DRONC coordinates cell death and compensatory proliferation.Mol Cell Biol. 2006 Oct;26(19):7258-68. doi: 10.1128/MCB.00183-06. Mol Cell Biol. 2006. PMID: 16980627 Free PMC article.
-
Drosophila caspase transduces Shaggy/GSK-3beta kinase activity in neural precursor development.EMBO J. 2005 Nov 2;24(21):3793-806. doi: 10.1038/sj.emboj.7600822. Epub 2005 Oct 13. EMBO J. 2005. PMID: 16222340 Free PMC article.
-
Jafrac2 is an IAP antagonist that promotes cell death by liberating Dronc from DIAP1.EMBO J. 2002 Oct 1;21(19):5118-29. doi: 10.1093/emboj/cdf530. EMBO J. 2002. PMID: 12356728 Free PMC article.
References
-
- Yuan J, Shaham S, Ledoux S, Ellis H M, Horvitz H R. Cell. 1993;75:641–652. - PubMed
-
- Yuan J, Horvitz H R. Development. 1992;116:309–320. - PubMed
-
- Hengartner M O, Ellis R E, Horvitz H R. Nature (London) 1992;356:494–499. - PubMed
-
- Ellis R E, Yuan J Y, Horvitz H R. Annu Rev Cell Biol. 1991;7:663–698. - PubMed
-
- Shaham S, Horvitz H R. Cell. 1996;86:201–208. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

