Characterization of functionally active subribosomal particles from Thermus aquaticus
- PMID: 9874776
- PMCID: PMC15097
- DOI: 10.1073/pnas.96.1.85
Characterization of functionally active subribosomal particles from Thermus aquaticus
Abstract
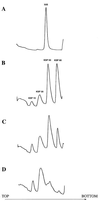
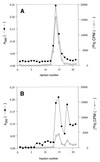
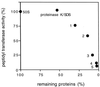
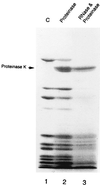
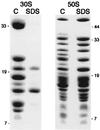
Peptidyl transferase activity of Thermus aquaticus ribosomes is resistant to the removal of a significant number of ribosomal proteins by protease digestion, SDS, and phenol extraction. To define the upper limit for the number of macromolecular components required for peptidyl transferase, particles obtained by extraction of T. aquaticus large ribosomal subunits were isolated and their RNA and protein composition was characterized. Active subribosomal particles contained both 23S and 5S rRNA associated with notable amounts of eight ribosomal proteins. N-terminal sequencing of the proteins identified them as L2, L3, L13, L15, L17, L18, L21, and L22. Ribosomal protein L4, which previously was thought to be essential for the reconstitution of particles active in peptide bond formation, was not found. These findings, together with the results of previous reconstitution experiments, reduce the number of possible essential macromolecular components of the peptidyl transferase center to 23S rRNA and ribosomal proteins L2 and L3. Complete removal of ribosomal proteins from T. aquaticus rRNA resulted in loss of tertiary folding of the particles and inactivation of peptidyl transferase. The accessibility of proteins in active subribosomal particles to proteinase hydrolysis was increased significantly after RNase treatment. These results and the observation that 50S ribosomal subunits exhibited much higher resistance to SDS extraction than 30S subunits are compatible with a proposed structural organization of the 50S subunit involving an RNA "cage" surrounding a core of a subset of ribosomal proteins.
Figures



Similar articles
-
Reconstitution of functionally active Thermus aquaticus large ribosomal subunits with in vitro-transcribed rRNA.Biochemistry. 1999 Feb 9;38(6):1780-8. doi: 10.1021/bi9822473. Biochemistry. 1999. PMID: 10026258
-
Effect of antibiotics on large ribosomal subunit assembly reveals possible function of 5 S rRNA.J Mol Biol. 1999 Sep 3;291(5):1025-34. doi: 10.1006/jmbi.1999.3030. J Mol Biol. 1999. PMID: 10518940
-
Unusual resistance of peptidyl transferase to protein extraction procedures.Science. 1992 Jun 5;256(5062):1416-9. doi: 10.1126/science.1604315. Science. 1992. PMID: 1604315
-
Histidine 229 in protein L2 is apparently essential for 50S peptidyl transferase activity.Biochem Cell Biol. 1995 Nov-Dec;73(11-12):1087-94. doi: 10.1139/o95-117. Biochem Cell Biol. 1995. PMID: 8722025 Review.
-
Mechanism of ribosome assisted protein folding: a new insight into rRNA functions.Biochem Biophys Res Commun. 2009 Jun 26;384(2):137-40. doi: 10.1016/j.bbrc.2009.04.106. Epub 2009 May 3. Biochem Biophys Res Commun. 2009. PMID: 19401192 Review.
Cited by
-
Magnesium Suppresses Defects in the Formation of 70S Ribosomes as Well as in Sporulation Caused by Lack of Several Individual Ribosomal Proteins.J Bacteriol. 2018 Aug 24;200(18):e00212-18. doi: 10.1128/JB.00212-18. Print 2018 Sep 15. J Bacteriol. 2018. PMID: 29967120 Free PMC article.
-
Comprehensive analysis of phosphorylated proteins of Escherichia coli ribosomes.J Proteome Res. 2009 Jul;8(7):3390-402. doi: 10.1021/pr900042e. J Proteome Res. 2009. PMID: 19469554 Free PMC article.
-
The Proto-Ribosome: an ancient nano-machine for peptide bond formation.Isr J Chem. 2010 Jun 18;50(1):29-35. doi: 10.1002/ijch.201000012. Isr J Chem. 2010. PMID: 26207070 Free PMC article.
-
Analysis of r-protein and RNA conformation of 30S subunit intermediates in bacteria.RNA. 2015 Jul;21(7):1323-34. doi: 10.1261/rna.048918.114. Epub 2015 May 21. RNA. 2015. PMID: 25999315 Free PMC article.
-
Functional genetic selection of Helix 66 in Escherichia coli 23S rRNA identified the eukaryotic-binding sequence for ribosomal protein L2.Nucleic Acids Res. 2007;35(12):4018-29. doi: 10.1093/nar/gkm356. Epub 2007 Jun 6. Nucleic Acids Res. 2007. PMID: 17553838 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

