Cytosolic adenylyl cyclase defines a unique signaling molecule in mammals
- PMID: 9874775
- PMCID: PMC15096
- DOI: 10.1073/pnas.96.1.79
Cytosolic adenylyl cyclase defines a unique signaling molecule in mammals
Abstract
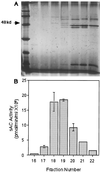
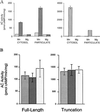
Mammals have nine differentially regulated isoforms of G protein-responsive transmembrane-spanning adenylyl cyclases. We now describe the existence of a distinct class of mammalian adenylyl cyclase that is soluble and insensitive to G protein or Forskolin regulation. Northern analysis indicates the gene encoding soluble adenylyl cyclase (sAC) is preferentially expressed in testis. As purified from rat testis cytosol, the active form of sAC appears to be a fragment derived from the full-length protein, suggesting a proteolytic mechanism for sAC activation. The two presumptive catalytic domains of sAC are closely related to cyanobacterial adenylyl cyclases, providing an evolutionary link between bacterial and mammalian signaling molecules.
Figures


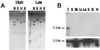
Similar articles
-
Identification and functional analysis of splice variants of the germ cell soluble adenylyl cyclase.J Biol Chem. 2001 Aug 24;276(34):31698-708. doi: 10.1074/jbc.M011698200. Epub 2001 Jun 21. J Biol Chem. 2001. PMID: 11423534
-
Structural analysis of human soluble adenylyl cyclase and crystal structures of its nucleotide complexes-implications for cyclase catalysis and evolution.FEBS J. 2014 Sep;281(18):4151-64. doi: 10.1111/febs.12913. Epub 2014 Jul 28. FEBS J. 2014. PMID: 25040695
-
Interaction of the two cytosolic domains of mammalian adenylyl cyclase.Proc Natl Acad Sci U S A. 1996 Jun 25;93(13):6621-5. doi: 10.1073/pnas.93.13.6621. Proc Natl Acad Sci U S A. 1996. PMID: 8692867 Free PMC article.
-
Bicarbonate-regulated soluble adenylyl cyclase.JOP. 2001 Jul;2(4 Suppl):154-8. JOP. 2001. PMID: 11875252 Review.
-
The class III adenylyl cyclases: multi-purpose signalling modules.Cell Signal. 2003 Dec;15(12):1081-9. doi: 10.1016/s0898-6568(03)00130-x. Cell Signal. 2003. PMID: 14575863 Review.
Cited by
-
A nuclear cAMP microdomain suppresses tumor growth by Hippo pathway inactivation.Cell Rep. 2022 Sep 27;40(13):111412. doi: 10.1016/j.celrep.2022.111412. Cell Rep. 2022. PMID: 36170819 Free PMC article.
-
High adenylyl cyclase activity and in vivo cAMP fluctuations in corals suggest central physiological role.Sci Rep. 2013;3:1379. doi: 10.1038/srep01379. Sci Rep. 2013. PMID: 23459251 Free PMC article.
-
Novel approach for the detection of the vestiges of testicular mRNA splicing errors in mature spermatozoa of Japanese Black bulls.PLoS One. 2013;8(2):e57296. doi: 10.1371/journal.pone.0057296. Epub 2013 Feb 26. PLoS One. 2013. PMID: 23468960 Free PMC article.
-
Self-renewal and differentiation of mouse embryonic stem cells as measured by Oct4 expression: the role of the cAMP/PKA pathway.In Vitro Cell Dev Biol Anim. 2007 Jan;43(1):37-47. doi: 10.1007/s11626-006-9001-5. In Vitro Cell Dev Biol Anim. 2007. PMID: 17570033
-
cAMP acts as a second messenger in pollen tube growth and reorientation.Proc Natl Acad Sci U S A. 2001 Aug 28;98(18):10481-6. doi: 10.1073/pnas.171104598. Epub 2001 Aug 21. Proc Natl Acad Sci U S A. 2001. PMID: 11517303 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

