Flow cytometric analysis of the in situ accessibility of Escherichia coli 16S rRNA for fluorescently labeled oligonucleotide probes
- PMID: 9835591
- PMCID: PMC90951
- DOI: 10.1128/AEM.64.12.4973-4982.1998
Flow cytometric analysis of the in situ accessibility of Escherichia coli 16S rRNA for fluorescently labeled oligonucleotide probes
Abstract
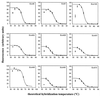
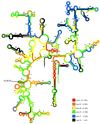
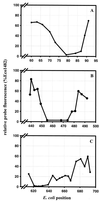
In situ identification of whole fixed bacterial cells by hybridization with fluorescently labeled, rRNA-targeted oligonucleotide probes is often limited by low signal intensities. In addition to an impermeability of the cell periphery and a low cellular rRNA content, the three-dimensional structure of the ribosome may hinder the access of oligonucleotides to their target sites. Until now, a systematic study on the accessibility of 16S rRNA target sites had not been done. Here, we report fluorescence intensities obtained with more than 200 oligonucleotide probes (mostly 18-mers) used with whole fixed cells of Escherichia coli DSM 30083(T). Two overlapping sets of adjacent oligonucleotides, 171 in total, were designed to cover the full length of the 16S rRNA. The two sets are shifted by 5 to 13 nucleotides. The probes were labeled with carboxyfluorescein, and signal intensities of hybridized cells were quantified by flow cytometry. Care was taken that the signal intensity of cells was dependent solely on the in situ accessibility of probe target sites. The brightest signal resulted from probe Eco1482, complementary to positions 1482 to 1499. With this probe, the fluorescence was 1.7 times brighter than that of the standard bacterial probe EUB338 and 44 times brighter than that of the worst probe, Eco468. The distribution of probe-conferred cell fluorescence in six arbitrarily set brightness classes (classes I to VI; 100 to 81%, 80 to 61%, 60 to 41%, 40 to 21%, 20 to 6%, and 5 to 0% of the brightness with Eco1482, respectively) was as follows: I, 4%; II, 14%; III, 21%; IV, 29%, V, 19%; and VI, 13%. A more detailed analysis of helices 6, 18, and 23 with additional probes demonstrated that a shift of the target region by only a few bases could result in a decline of cell fluorescence from >80 to <10%. Considering the high evolutionary conservation of 16S rRNA, the in situ accessibility map of E. coli should facilitate a more rational selection of probe target sites for other species as well.
Figures
Similar articles
-
In situ accessibility of Escherichia coli 23S rRNA to fluorescently labeled oligonucleotide probes.Appl Environ Microbiol. 2001 Feb;67(2):961-8. doi: 10.1128/AEM.67.2.961-968.2001. Appl Environ Microbiol. 2001. PMID: 11157269 Free PMC article.
-
Making all parts of the 16S rRNA of Escherichia coli accessible in situ to single DNA oligonucleotides.Appl Environ Microbiol. 2006 Jan;72(1):733-44. doi: 10.1128/AEM.72.1.733-744.2006. Appl Environ Microbiol. 2006. PMID: 16391113 Free PMC article.
-
In situ accessibility of small-subunit rRNA of members of the domains Bacteria, Archaea, and Eucarya to Cy3-labeled oligonucleotide probes.Appl Environ Microbiol. 2003 Mar;69(3):1748-58. doi: 10.1128/AEM.69.3.1748-1758.2003. Appl Environ Microbiol. 2003. PMID: 12620867 Free PMC article.
-
Double labeling of oligonucleotide probes for fluorescence in situ hybridization (DOPE-FISH) improves signal intensity and increases rRNA accessibility.Appl Environ Microbiol. 2010 Feb;76(3):922-6. doi: 10.1128/AEM.02456-09. Epub 2009 Dec 4. Appl Environ Microbiol. 2010. PMID: 19966029 Free PMC article.
-
ProBac-seq, a bacterial single-cell RNA sequencing methodology using droplet microfluidics and large oligonucleotide probe sets.Nat Protoc. 2024 Oct;19(10):2939-2966. doi: 10.1038/s41596-024-01002-1. Epub 2024 May 20. Nat Protoc. 2024. PMID: 38769144 Review.
Cited by
-
Fluorescent In Situ hybridization: a new tool for the direct identification and detection of F. psychrophilum.PLoS One. 2012;7(11):e49280. doi: 10.1371/journal.pone.0049280. Epub 2012 Nov 9. PLoS One. 2012. PMID: 23152887 Free PMC article.
-
Molecular monitoring of ruminal prevotellas.Folia Microbiol (Praha). 2001;46(1):87-90. doi: 10.1007/BF02825895. Folia Microbiol (Praha). 2001. PMID: 11501487
-
Utilization of tmRNA sequences for bacterial identification.BMC Microbiol. 2001 Sep 7;1:20. doi: 10.1186/1471-2180-1-20. BMC Microbiol. 2001. PMID: 11560762 Free PMC article.
-
Comparative 16S rRNA analysis of lake bacterioplankton reveals globally distributed phylogenetic clusters including an abundant group of actinobacteria.Appl Environ Microbiol. 2000 Nov;66(11):5053-65. doi: 10.1128/AEM.66.11.5053-5065.2000. Appl Environ Microbiol. 2000. PMID: 11055963 Free PMC article.
-
In situ accessibility of Saccharomyces cerevisiae 26S rRNA to Cy3-labeled oligonucleotide probes comprising the D1 and D2 domains.Appl Environ Microbiol. 2003 May;69(5):2899-905. doi: 10.1128/AEM.69.5.2899-2905.2003. Appl Environ Microbiol. 2003. PMID: 12732564 Free PMC article.
References
-
- Amann R, Ludwig W, Schulze R, Spring S, Moore E, Schleifer K H. rRNA-targeted oligonucleotide probes for the identification of genuine and former pseudomonads. Syst Appl Microbiol. 1996;19:501–509.
-
- Amann, R. Unpublished results.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

