ASAP1, a phospholipid-dependent arf GTPase-activating protein that associates with and is phosphorylated by Src
- PMID: 9819391
- PMCID: PMC109286
- DOI: 10.1128/MCB.18.12.7038
ASAP1, a phospholipid-dependent arf GTPase-activating protein that associates with and is phosphorylated by Src
Abstract
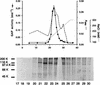
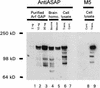
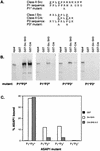
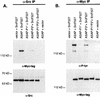
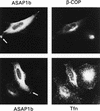
Membrane trafficking is regulated in part by small GTP-binding proteins of the ADP-ribosylation factor (Arf) family. Arf function depends on the controlled exchange and hydrolysis of GTP. We have purified and cloned two variants of a 130-kDa phosphatidylinositol 4, 5-biphosphate (PIP2)-dependent Arf1 GTPase-activating protein (GAP), which we call ASAP1a and ASAP1b. Both contain a pleckstrin homology (PH) domain, a zinc finger similar to that found in another Arf GAP, three ankyrin (ANK) repeats, a proline-rich region with alternative splicing and SH3 binding motifs, eight repeats of the sequence E/DLPPKP, and an SH3 domain. Together, the PH, zinc finger, and ANK repeat regions possess PIP2-dependent GAP activity on Arf1 and Arf5, less activity on Arf6, and no detectable activity on Arl2 in vitro. The cDNA for ASAP1 was independently identified in a screen for proteins that interact with the SH3 domain of the tyrosine kinase Src. ASAP1 associates in vitro with the SH3 domains of Src family members and with the Crk adapter protein. ASAP1 coprecipitates with Src from cell lysates and is phosphorylated on tyrosine residues in cells expressing activated Src. Both coimmunoprecipitation and tyrosine phosphorylation depend on the same proline-rich class II Src SH3 binding site required for in vitro association. By directly interacting with both Arfs and tyrosine kinases involved in regulating cell growth and cytoskeletal organization, ASAP1 could coordinate membrane remodeling events with these processes.
Figures






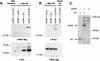
Similar articles
-
Interaction of the N terminus of ADP-ribosylation factor with the PH domain of the GTPase-activating protein ASAP1 requires phosphatidylinositol 4,5-bisphosphate.J Biol Chem. 2019 Nov 15;294(46):17354-17370. doi: 10.1074/jbc.RA119.009269. Epub 2019 Oct 6. J Biol Chem. 2019. PMID: 31591270 Free PMC article.
-
Mislocalization or reduced expression of Arf GTPase-activating protein ASAP1 inhibits cell spreading and migration by influencing Arf1 GTPase cycling.J Biol Chem. 2005 Mar 11;280(10):8884-92. doi: 10.1074/jbc.M412200200. Epub 2005 Jan 4. J Biol Chem. 2005. PMID: 15632162
-
ACAPs are arf6 GTPase-activating proteins that function in the cell periphery.J Cell Biol. 2000 Oct 30;151(3):627-38. doi: 10.1083/jcb.151.3.627. J Cell Biol. 2000. PMID: 11062263 Free PMC article.
-
Activation of toxin ADP-ribosyltransferases by the family of ADP-ribosylation factors.Adv Exp Med Biol. 1997;419:315-20. doi: 10.1007/978-1-4419-8632-0_41. Adv Exp Med Biol. 1997. PMID: 9193671 Review.
-
Activation of toxin ADP-ribosyltransferases by eukaryotic ADP-ribosylation factors.Mol Cell Biochem. 1999 Mar;193(1-2):153-7. Mol Cell Biochem. 1999. PMID: 10331652 Review.
Cited by
-
CIN85 associates with multiple effectors controlling intracellular trafficking of epidermal growth factor receptors.Mol Biol Cell. 2004 Jul;15(7):3155-66. doi: 10.1091/mbc.e03-09-0683. Epub 2004 Apr 16. Mol Biol Cell. 2004. PMID: 15090612 Free PMC article.
-
A new paxillin-binding protein, PAG3/Papalpha/KIAA0400, bearing an ADP-ribosylation factor GTPase-activating protein activity, is involved in paxillin recruitment to focal adhesions and cell migration.Mol Biol Cell. 2000 Apr;11(4):1315-27. doi: 10.1091/mbc.11.4.1315. Mol Biol Cell. 2000. PMID: 10749932 Free PMC article.
-
Paxillin-kinase-linker tyrosine phosphorylation regulates directional cell migration.Mol Biol Cell. 2009 Nov;20(22):4706-19. doi: 10.1091/mbc.e09-07-0548. Epub 2009 Sep 23. Mol Biol Cell. 2009. PMID: 19776348 Free PMC article.
-
Regulation of SRC family kinases in human cancers.J Signal Transduct. 2011;2011:865819. doi: 10.1155/2011/865819. Epub 2011 Apr 4. J Signal Transduct. 2011. PMID: 21776389 Free PMC article.
-
Membrane surface recognition by the ASAP1 PH domain and consequences for interactions with the small GTPase Arf1.Sci Adv. 2020 Sep 30;6(40):eabd1882. doi: 10.1126/sciadv.abd1882. Print 2020 Sep. Sci Adv. 2020. PMID: 32998886 Free PMC article.
References
-
- Alexandropoulos K, Baltimore D. Coordinate activation of c-Src by SH3- and SH2-binding sites on a novel p130Cas-related protein, Sin. Genes Dev. 1996;10:1341–1355. - PubMed
-
- Allen C M, Ely C M, Juaneza M A, Parsons S J. Activation of Fyn tyrosine kinase upon secretogogue stimulation of bovine chromaffin cells. J Neurosci Res. 1996;44:421–429. - PubMed
-
- Antonny B, Huber I, Paris S, Chabre M, Cassel D. Activation of ADP-ribosylation factor 1 GTPase-activating protein by phosphatidylcholine-derived diacylglycerols. J Biol Chem. 1997;272:30848–30851. - PubMed
-
- Barnekow A, Jahn R, Schartl M. Synaptophysin: a substrate for the protein tyrosine kinase pp60c-src in intact synaptic vesicles. Oncogene. 1990;5:1019–1024. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
