In vivo function of Hsp90 is dependent on ATP binding and ATP hydrolysis
- PMID: 9817749
- PMCID: PMC2132952
- DOI: 10.1083/jcb.143.4.901
In vivo function of Hsp90 is dependent on ATP binding and ATP hydrolysis
Abstract
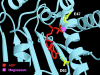
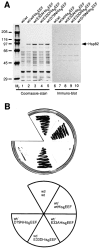
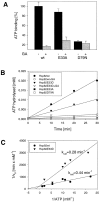
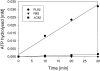
Heat shock protein 90 (Hsp90), an abundant molecular chaperone in the eukaryotic cytosol, is involved in the folding of a set of cell regulatory proteins and in the re-folding of stress-denatured polypeptides. The basic mechanism of action of Hsp90 is not yet understood. In particular, it has been debated whether Hsp90 function is ATP dependent. A recent crystal structure of the NH2-terminal domain of yeast Hsp90 established the presence of a conserved nucleotide binding site that is identical with the binding site of geldanamycin, a specific inhibitor of Hsp90. The functional significance of nucleotide binding by Hsp90 has remained unclear. Here we present evidence for a slow but clearly detectable ATPase activity in purified Hsp90. Based on a new crystal structure of the NH2-terminal domain of human Hsp90 with bound ADP-Mg and on the structural homology of this domain with the ATPase domain of Escherichia coli DNA gyrase, the residues of Hsp90 critical in ATP binding (D93) and ATP hydrolysis (E47) were identified. The corresponding mutations were made in the yeast Hsp90 homologue, Hsp82, and tested for their ability to functionally replace wild-type Hsp82. Our results show that both ATP binding and hydrolysis are required for Hsp82 function in vivo. The mutant Hsp90 proteins tested are defective in the binding and ATP hydrolysis-dependent cycling of the co-chaperone p23, which is thought to regulate the binding and release of substrate polypeptide from Hsp90. Remarkably, the complete Hsp90 protein is required for ATPase activity and for the interaction with p23, suggesting an intricate allosteric communication between the domains of the Hsp90 dimer. Our results establish Hsp90 as an ATP-dependent chaperone.
Figures



Similar articles
-
Polypeptide release by Hsp90 involves ATP hydrolysis and is enhanced by the co-chaperone p23.EMBO J. 2000 Nov 1;19(21):5930-40. doi: 10.1093/emboj/19.21.5930. EMBO J. 2000. PMID: 11060043 Free PMC article.
-
Identification and structural characterization of the ATP/ADP-binding site in the Hsp90 molecular chaperone.Cell. 1997 Jul 11;90(1):65-75. doi: 10.1016/s0092-8674(00)80314-1. Cell. 1997. PMID: 9230303
-
C-terminal regions of Hsp90 are important for trapping the nucleotide during the ATPase cycle.J Mol Biol. 2000 Nov 3;303(4):583-92. doi: 10.1006/jmbi.2000.4157. J Mol Biol. 2000. PMID: 11054293
-
Novobiocin and additional inhibitors of the Hsp90 C-terminal nucleotide-binding pocket.Curr Med Chem. 2008;15(26):2702-17. doi: 10.2174/092986708786242895. Curr Med Chem. 2008. PMID: 18991631 Free PMC article. Review.
-
[Molecular chaperone HSP90 as a novel target for cancer chemotherapy].Nihon Yakurigaku Zasshi. 2003 Jan;121(1):33-42. doi: 10.1254/fpj.121.33. Nihon Yakurigaku Zasshi. 2003. PMID: 12617036 Review. Japanese.
Cited by
-
BRI1 and BAK1 Canonical Distribution in Plasma Membrane Is HSP90 Dependent.Cells. 2022 Oct 22;11(21):3341. doi: 10.3390/cells11213341. Cells. 2022. PMID: 36359737 Free PMC article.
-
The ATPase cycle of Hsp90 drives a molecular 'clamp' via transient dimerization of the N-terminal domains.EMBO J. 2000 Aug 15;19(16):4383-92. doi: 10.1093/emboj/19.16.4383. EMBO J. 2000. PMID: 10944121 Free PMC article.
-
Nucleotide-dependent interaction of Saccharomyces cerevisiae Hsp90 with the cochaperone proteins Sti1, Cpr6, and Sba1.Mol Cell Biol. 2007 Jan;27(2):768-76. doi: 10.1128/MCB.01034-06. Epub 2006 Nov 13. Mol Cell Biol. 2007. PMID: 17101799 Free PMC article.
-
Hsp90 selectively modulates phenotype in vertebrate development.PLoS Genet. 2007 Mar 30;3(3):e43. doi: 10.1371/journal.pgen.0030043. Epub 2007 Feb 8. PLoS Genet. 2007. PMID: 17397257 Free PMC article.
-
A transmembrane guanylyl cyclase (DAF-11) and Hsp90 (DAF-21) regulate a common set of chemosensory behaviors in caenorhabditis elegans.Genetics. 2000 May;155(1):85-104. doi: 10.1093/genetics/155.1.85. Genetics. 2000. PMID: 10790386 Free PMC article.
References
-
- Ausubel, F.M., R. Brent, R.E. Kingston, D.D. Moore, J.G. Seidman, J.A. Smith, and K. Struhl. 1987. Current Protocols in Molecular Biology. Wiley and Sons, Inc., New York. 8.5.1–8.5.9.
-
- Bartel, P.L., C.-T. Chien, R. Sternglanz, and S. Fields. 1993. Using the two-hybrid system to detect protein–protein interactions. In Cellular Interactions in Development: A Practical Approach. D.A. Hartley, editor. Oxford University Press, Oxford. 153–179.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

