Formation and amplification of a novel tombusvirus defective RNA which lacks the 5' nontranslated region of the viral genome
- PMID: 9811726
- PMCID: PMC110502
- DOI: 10.1128/JVI.72.12.9897-9905.1998
Formation and amplification of a novel tombusvirus defective RNA which lacks the 5' nontranslated region of the viral genome
Abstract
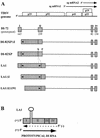
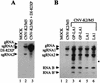
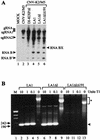
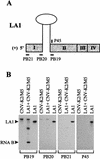
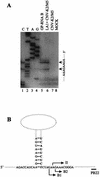
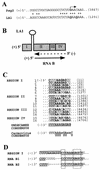
Defective interfering (DI) RNAs of tomato bushy stunt virus (TBSV) are small, subgenomic, helper-dependent replicons that are believed to be generated primarily by aberrant events during replication of the plus-sense RNA genome. Prototypical TBSV DI RNAs contain four noncontiguous segments (regions I through IV) derived from the 5' nontranslated region (NTR) (I), an internal section (II), and the 3'-terminal portion (III and IV) of the viral genome. We have studied the formation of these molecules by using engineered precursor DI RNA transcripts and report here the consistent accumulation of a novel defective RNA species, designated RNA B. Northern blot, primer extension, and sequence analyses indicated that, unlike prototypical DI RNAs, RNA B lacks region I. In vitro transcripts corresponding to the region II-III-IV structure of RNA B were amplified when coinoculated with helper, indicating that the 5' NTR of the genome does not harbor cis-acting replication elements essential for viral RNA replication. Region I is, however, important for DI RNA fitness, since molecules lacking it accumulated to significantly lower levels ( approximately 10-fold reduction). Analysis of the minus-strand sequence of region I led to the identification of an RNA undecamer sequence, arranged in tandem, at its very 3' terminus. Additional variants of the undecamer motif were also identified at internal positions in region I and in the negative strands of regions II, III, and IV. Features of the undecamer motif, the consensus of which is (-)3'-CCCAAAGAGAG, are consistent with a role as a cis-acting replication element. It is proposed that the ability of RNA B to be amplified is due, in part, to compensatory effects of a strategically positioned undecamer motif in region II. Possible replicase-mediated mechanisms for the generation of this novel viral RNA are also presented.
Figures

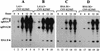
Similar articles
-
Enhancer-like properties of an RNA element that modulates Tombusvirus RNA accumulation.Virology. 1999 Mar 30;256(1):162-71. doi: 10.1006/viro.1999.9630. Virology. 1999. PMID: 10087236
-
Nonhomologous RNA recombination in tombusviruses: generation and evolution of defective interfering RNAs by stepwise deletions.J Virol. 1994 Jan;68(1):14-24. doi: 10.1128/JVI.68.1.14-24.1994. J Virol. 1994. PMID: 8254723 Free PMC article.
-
An RNA domain within the 5' untranslated region of the tomato bushy stunt virus genome modulates viral RNA replication.J Mol Biol. 2001 Jan 26;305(4):741-56. doi: 10.1006/jmbi.2000.4298. J Mol Biol. 2001. PMID: 11162089
-
Tombusvirus polymerase: Structure and function.Virus Res. 2017 Apr 15;234:74-86. doi: 10.1016/j.virusres.2017.01.012. Epub 2017 Jan 19. Virus Res. 2017. PMID: 28111194 Review.
-
Intragenomic Long-Distance RNA-RNA Interactions in Plus-Strand RNA Plant Viruses.Front Microbiol. 2018 Apr 4;9:529. doi: 10.3389/fmicb.2018.00529. eCollection 2018. Front Microbiol. 2018. PMID: 29670583 Free PMC article. Review.
Cited by
-
Defective RNA Particles of Plant Viruses-Origin, Structure and Role in Pathogenesis.Viruses. 2022 Dec 16;14(12):2814. doi: 10.3390/v14122814. Viruses. 2022. PMID: 36560818 Free PMC article. Review.
-
Defective Interfering RNAs: Foes of Viruses and Friends of Virologists.Viruses. 2009 Dec;1(3):895-919. doi: 10.3390/v1030895. Epub 2009 Nov 10. Viruses. 2009. PMID: 21994575 Free PMC article.
-
The p92 polymerase coding region contains an internal RNA element required at an early step in Tombusvirus genome replication.J Virol. 2005 Apr;79(8):4848-58. doi: 10.1128/JVI.79.8.4848-4858.2005. J Virol. 2005. PMID: 15795270 Free PMC article.
-
Structural properties of a multifunctional T-shaped RNA domain that mediate efficient tomato bushy stunt virus RNA replication.J Virol. 2004 Oct;78(19):10490-500. doi: 10.1128/JVI.78.19.10490-10500.2004. J Virol. 2004. PMID: 15367615 Free PMC article.
-
A complex network of RNA-RNA interactions controls subgenomic mRNA transcription in a tombusvirus.EMBO J. 2004 Aug 18;23(16):3365-74. doi: 10.1038/sj.emboj.7600336. Epub 2004 Jul 29. EMBO J. 2004. PMID: 15282544 Free PMC article.
References
-
- Burgyan J, Rubino L, Russo M. De novo generation of cymbidium ringspot virus defective interfering RNA. J Gen Virol. 1991;72:505–509. - PubMed
-
- Celix A, Rodriguez-Cerezo E, Garcia-Arenal F. New satellite RNAs, but not DI RNAs, are found in natural populations of tomato bushy stunt virus. Virology. 1997;239:277–284. - PubMed
-
- Chang Y C, Borja M, Scholthof H B, Jackson A O, Morris T J. Host effects and sequences essential for accumulation of defective interfering RNAs of cucumber necrosis and tomato bushy stunt tombusviruses. Virology. 1995;210:41–53. - PubMed
-
- Dreher T W, Hall T C. Mutational analysis of the sequence and structural requirements in brome mosaic virus RNA for minus strand promoter activity. J Mol Biol. 1988;210:31–40. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

