The neuronal RNA-binding protein Nova-2 is implicated as the autoantigen targeted in POMA patients with dementia
- PMID: 9789075
- PMCID: PMC23773
- DOI: 10.1073/pnas.95.22.13254
The neuronal RNA-binding protein Nova-2 is implicated as the autoantigen targeted in POMA patients with dementia
Abstract
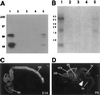
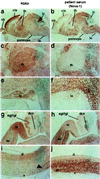
Paraneoplastic opsoclonus myoclonus ataxia (POMA) is a neurologic disorder thought to be mediated by an autoimmune attack against onconeural disease antigens that are expressed by gynecologic or lung tumors and by neurons. One POMA disease antigen, termed Nova-1, has been identified as a neuron-specific KH-type RNA-binding protein. Nova-1 expression is restricted to specific regions of the central nervous system, primarily the hindbrain and ventral spinal cord, which correlate with the predominantly motor symptoms in POMA. However, POMA antisera recognize antigens that are widely expressed in both caudal and rostral regions of the central nervous system, and some patients develop cognitive symptoms. We have used POMA antisera to clone a cDNA encoding a second POMA disease antigen termed Nova-2. Nova-2 is closely related to Nova-1, and is expressed at high levels in neurons during development and in adulthood, and at lower levels in the adult lung. In the postnatal mouse brain, Nova-2 is expressed in a pattern that is largely reciprocal with Nova-1, including high levels of Nova-2 expression in the neocortex and hippocampus. Functional characterization of Nova-2 in RNA selection and nitrocellulose filter-binding assays reveals that Nova-2 binds RNA with high affinity and with sequence specificity that differs from Nova-1. Our results demonstrate that the immune response in POMA targets a family of highly related sequence-specific neuronal RNA-binding proteins. The expression pattern of the Nova-2 protein is likely to underlie the development of cognitive deficits in some POMA patients.
Figures


Similar articles
-
The onconeural antigen Nova-1 is a neuron-specific RNA-binding protein, the activity of which is inhibited by paraneoplastic antibodies.J Neurosci. 1996 Feb 1;16(3):1114-22. doi: 10.1523/JNEUROSCI.16-03-01114.1996. J Neurosci. 1996. PMID: 8558240 Free PMC article.
-
Nova, the paraneoplastic Ri antigen, is homologous to an RNA-binding protein and is specifically expressed in the developing motor system.Neuron. 1993 Oct;11(4):657-72. doi: 10.1016/0896-6273(93)90077-5. Neuron. 1993. PMID: 8398153
-
The neuronal RNA binding protein Nova-1 recognizes specific RNA targets in vitro and in vivo.Mol Cell Biol. 1997 Jun;17(6):3194-201. doi: 10.1128/MCB.17.6.3194. Mol Cell Biol. 1997. PMID: 9154818 Free PMC article.
-
Autoimmune central nervous system paraneoplastic disorders: mechanisms, diagnosis, and therapeutic options.Ann Neurol. 1995 May;37 Suppl 1:S102-13. doi: 10.1002/ana.410370711. Ann Neurol. 1995. PMID: 8968221 Review.
-
Developing global insight into RNA regulation.Cold Spring Harb Symp Quant Biol. 2006;71:321-7. doi: 10.1101/sqb.2006.71.002. Cold Spring Harb Symp Quant Biol. 2006. PMID: 17381312 Review.
Cited by
-
The Genetic Control of Stoichiometry Underlying Autism.Annu Rev Neurosci. 2020 Jul 8;43:509-533. doi: 10.1146/annurev-neuro-100119-024851. Annu Rev Neurosci. 2020. PMID: 32640929 Free PMC article.
-
NOVA-dependent regulation of cryptic NMD exons controls synaptic protein levels after seizure.Elife. 2013 Jan 22;2:e00178. doi: 10.7554/eLife.00178. Elife. 2013. PMID: 23359859 Free PMC article.
-
Autoimmune Encephalitis With Autoimmune Diabetes: A Case of Horror Autotoxicus.Cureus. 2023 Jan 27;15(1):e34268. doi: 10.7759/cureus.34268. eCollection 2023 Jan. Cureus. 2023. PMID: 36855486 Free PMC article.
-
Differential NOVA2-Mediated Splicing in Excitatory and Inhibitory Neurons Regulates Cortical Development and Cerebellar Function.Neuron. 2019 Feb 20;101(4):707-720.e5. doi: 10.1016/j.neuron.2018.12.019. Epub 2019 Jan 9. Neuron. 2019. PMID: 30638744 Free PMC article.
-
Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches.Nat Rev Mol Cell Biol. 2009 Nov;10(11):741-54. doi: 10.1038/nrm2777. Epub 2009 Sep 23. Nat Rev Mol Cell Biol. 2009. PMID: 19773805 Free PMC article. Review.
References
-
- Posner J B, Furneaux H M. In: Immunologic Mechanisms in Neurologic and Psychiatric Disease. Waksman B H, editor. New York: Raven; 1990. pp. 187–217.
-
- Gray-Keller M P, Polans A S, Palczewski K, Detwiler P B. Neuron. 1993;10:523–531. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

