Residues in the Swi5 zinc finger protein that mediate cooperative DNA binding with the Pho2 homeodomain protein
- PMID: 9774660
- PMCID: PMC109230
- DOI: 10.1128/MCB.18.11.6436
Residues in the Swi5 zinc finger protein that mediate cooperative DNA binding with the Pho2 homeodomain protein
Abstract
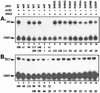
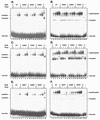
The Swi5 zinc finger and the Pho2 homeodomain DNA-binding proteins bind cooperatively to the HO promoter. Pho2 (also known as Bas2 or Grf10) activates transcription of diverse genes, acting with multiple distinct DNA-binding proteins. We have performed a genetic screen to identify amino acid residues in Swi5 that are required for synergistic transcriptional activation of a reporter construct in vivo. Nine unique amino acid substitutions within a 24-amino-acid region of Swi5, upstream of the DNA-binding domain, reduce expression of promoters that require both Swi5 and Pho2 for activation. In vitro DNA binding experiments show that the mutant Swi5 proteins bind DNA normally, but some mutant Swi5 proteins (resulting from SWI5* mutations) show reduced cooperative DNA binding with Pho2. In vivo experiments show that these SWI5* mutations sharply reduce expression of promoters that require both SWI5 and PHO2, while expression of promoters that require SWI5 but are PHO2 independent is largely unaffected. This suggests that these SWI5* mutations do not affect the ability of Swi5 to bind DNA or activate transcription but specifically affect the region of Swi5 required for interaction with Pho2. Two-hybrid experiments show that amino acids 471 to 513 of Swi5 are necessary and sufficient for interaction with Pho2 and that the SWI5* point mutations cause a severe reduction in this two-hybrid interaction. Analysis of promoter activation by these mutants suggests that this small region of Swi5 has at least two distinct functions, conferring specificity for activation of the HO promoter and for interaction with Pho2.
Figures




Similar articles
-
The Swi5 zinc-finger and Grf10 homeodomain proteins bind DNA cooperatively at the yeast HO promoter.Proc Natl Acad Sci U S A. 1993 Dec 1;90(23):11237-41. doi: 10.1073/pnas.90.23.11237. Proc Natl Acad Sci U S A. 1993. PMID: 7902583 Free PMC article.
-
Mutations in the pho2 (bas2) transcription factor that differentially affect activation with its partner proteins bas1, pho4, and swi5.J Biol Chem. 2002 Oct 4;277(40):37612-8. doi: 10.1074/jbc.M206125200. Epub 2002 Jul 26. J Biol Chem. 2002. PMID: 12145299
-
Identification and purification of a protein that binds DNA cooperatively with the yeast SWI5 protein.Mol Cell Biol. 1993 Sep;13(9):5524-37. doi: 10.1128/mcb.13.9.5524-5537.1993. Mol Cell Biol. 1993. PMID: 8355698 Free PMC article.
-
Long-range interactions at the HO promoter.Mol Cell Biol. 1997 May;17(5):2669-78. doi: 10.1128/MCB.17.5.2669. Mol Cell Biol. 1997. PMID: 9111337 Free PMC article.
-
Regulating the HO endonuclease in yeast.Curr Opin Genet Dev. 1993 Apr;3(2):286-94. doi: 10.1016/0959-437x(93)90036-o. Curr Opin Genet Dev. 1993. PMID: 8504254 Review.
Cited by
-
Disruption of yeast forkhead-associated cell cycle transcription by oxidative stress.Mol Biol Cell. 2004 Dec;15(12):5659-69. doi: 10.1091/mbc.e04-04-0340. Epub 2004 Sep 15. Mol Biol Cell. 2004. PMID: 15371544 Free PMC article.
-
Chd1 and yFACT act in opposition in regulating transcription.Mol Cell Biol. 2007 Sep;27(18):6279-87. doi: 10.1128/MCB.00978-07. Epub 2007 Jul 9. Mol Cell Biol. 2007. PMID: 17620414 Free PMC article.
-
SWI/SNF binding to the HO promoter requires histone acetylation and stimulates TATA-binding protein recruitment.Mol Cell Biol. 2006 Jun;26(11):4095-110. doi: 10.1128/MCB.01849-05. Mol Cell Biol. 2006. PMID: 16705163 Free PMC article.
-
Genetic interactions between Nhp6 and Gcn5 with Mot1 and the Ccr4-Not complex that regulate binding of TATA-binding protein in Saccharomyces cerevisiae.Genetics. 2006 Feb;172(2):837-49. doi: 10.1534/genetics.105.050245. Epub 2005 Nov 4. Genetics. 2006. PMID: 16272410 Free PMC article.
-
A scale of functional divergence for yeast duplicated genes revealed from analysis of the protein-protein interaction network.Genome Biol. 2004;5(10):R76. doi: 10.1186/gb-2004-5-10-r76. Epub 2004 Sep 15. Genome Biol. 2004. PMID: 15461795 Free PMC article.
References
-
- Ausubel F M, Brent R, Kingston R E, Moore D E, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1987.
-
- Bobola N, Jansen R P, Shin T H, Nasmyth K. Asymmetric accumulation of Ash1p in postanaphase nuclei depends on a myosin and restricts yeast mating-type switching to mother cells. Cell. 1996;84:699–709. - PubMed
-
- Brazas R M, Bhoite L T, Murphy M D, Yu Y, Chen Y, Neklason D W, Stillman D J. Determining the requirements for cooperative DNA binding by Swi5p and Pho2p (Grf10p/Bas2p) at the HO promoter. J Biol Chem. 1995;270:29151–29161. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
