Potential contributions of viral envelope and host genetic factors in a human immunodeficiency virus type 1-infected long-term survivor
- PMID: 9765405
- PMCID: PMC110277
- DOI: 10.1128/JVI.72.11.8650-8658.1998
Potential contributions of viral envelope and host genetic factors in a human immunodeficiency virus type 1-infected long-term survivor
Abstract
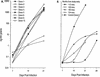
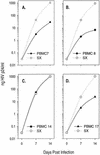
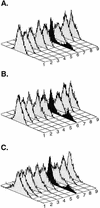
The lack of clinical progression in some individuals despite prolonged human immunodeficiency virus type 1 (HIV-1) infection may result from infection with less-pathogenic viral strains. To address this question, we examined the HIV-1 envelope protein from a donor with a low viral burden, stable CD4(+) T-lymphocyte counts, and little evidence of CD8(+) T-cell expansion, activation, or immune activity. To avoid potential changes in envelope function resulting from selection in vitro, envelope clones were constructed by using viral RNA isolated from uncultured peripheral blood mononuclear cells (PBMC). The data showed that recombinant viruses containing envelope sequences derived from RNA isolated from patient PBMC replicated poorly in primary CD4(+) T cells but demonstrated efficient growth in macrophages. The unusual phenotype of these viruses could not be explained solely by differential utilization of coreceptors since the chimeric viruses, as well as an uncloned isolate obtained from the same visit date, can utilize CCR5. In addition, the donor's own cells appeared resistant to infection with chimeric viruses containing autologous envelope sequences. Genotype analysis revealed that the donor was heterozygous for the previously described 32-bp deletion in CCR5 which may be linked with prolonged survival in HIV-1-infected individuals. These data suggest that the changes in envelope sequences confer properties of viral attenuation, which together with the CCR5 +/Delta32 genotype could account for the long-term survival of this patient.
Figures


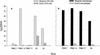

Similar articles
-
Generation and properties of a human immunodeficiency virus type 1 isolate resistant to the small molecule CCR5 inhibitor, SCH-417690 (SCH-D).Virology. 2005 Jul 20;338(1):182-99. doi: 10.1016/j.virol.2005.04.035. Virology. 2005. PMID: 15935415
-
Phenotype and envelope gene diversity of nef-deleted HIV-1 isolated from long-term survivors infected from a single source.Virol J. 2007 Jul 16;4:75. doi: 10.1186/1743-422X-4-75. Virol J. 2007. PMID: 17634131 Free PMC article.
-
Infection of baboons with a simian immunodeficiency virus/HIV-1 chimeric virus constructed with an HIV-1 Thai subtype E envelope.AIDS. 1998 May 28;12(8):849-57. doi: 10.1097/00002030-199808000-00006. AIDS. 1998. PMID: 9631137
-
Host and Viral Factors in HIV-Mediated Bystander Apoptosis.Viruses. 2017 Aug 22;9(8):237. doi: 10.3390/v9080237. Viruses. 2017. PMID: 28829402 Free PMC article. Review.
-
Virus-host interactions in HIV pathogenesis: directions for therapy.Adv Dent Res. 2011 Apr;23(1):13-8. doi: 10.1177/0022034511398874. Adv Dent Res. 2011. PMID: 21441474 Free PMC article. Review.
Cited by
-
Induction of humoral immune responses following vaccination with envelope-containing, formaldehyde-treated, thermally inactivated human immunodeficiency virus type 1.J Virol. 2005 Apr;79(8):4927-35. doi: 10.1128/JVI.79.8.4927-4935.2005. J Virol. 2005. PMID: 15795278 Free PMC article.
-
Formaldehyde-treated, heat-inactivated virions with increased human immunodeficiency virus type 1 env can be used to induce high-titer neutralizing antibody responses.J Virol. 2005 Aug;79(16):10210-7. doi: 10.1128/JVI.79.16.10210-10217.2005. J Virol. 2005. PMID: 16051814 Free PMC article.
-
Relationship between HIV coreceptor tropism and disease progression in persons with untreated chronic HIV infection.J Acquir Immune Defic Syndr. 2009 Mar 1;50(3):259-66. doi: 10.1097/QAI.0b013e3181989a8b. J Acquir Immune Defic Syndr. 2009. PMID: 19194318 Free PMC article.
-
Low human immunodeficiency virus envelope diversity correlates with low in vitro replication capacity and predicts spontaneous control of plasma viremia after treatment interruptions.J Virol. 2005 Jul;79(14):9026-37. doi: 10.1128/JVI.79.14.9026-9037.2005. J Virol. 2005. PMID: 15994796 Free PMC article.
-
HIV replication capacity is an independent predictor of disease progression in persons with untreated chronic HIV infection.J Acquir Immune Defic Syndr. 2010 Apr 1;53(4):472-9. doi: 10.1097/QAI.0b013e3181cae480. J Acquir Immune Defic Syndr. 2010. PMID: 20032783 Free PMC article.
References
-
- Alkhatib G, Combadiere C, Broder C C, Feng Y, Kennedy P E, Murphy P M, Berger E A. CC CKR5: a RANTES, MIP-1α, MIP-1β receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. - PubMed
-
- Baba T W, Jeong Y S, Pennick D, Bronson R, Greene M F, Ruprecht R M. Pathogenicity of live, attenuated SIV after mucosal infection of neonatal macaques. Science. 1995;267:1820–1825. - PubMed
-
- Balliet J W, Kolson D L, Eiger G, Kim F M, McGann K A, Srinivasan A, Collman R. Distinct effects in primary macrophages and lymphocytes of the human immunodeficiency virus type 1 accessory genes vpr, vpu, and nef: mutational analysis of a primary HIV-1 isolate. Virology. 1994;200:623–631. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

