Dynamics of the genome during early Xenopus laevis development: karyomeres as independent units of replication
- PMID: 9732278
- PMCID: PMC2149347
- DOI: 10.1083/jcb.142.5.1159
Dynamics of the genome during early Xenopus laevis development: karyomeres as independent units of replication
Abstract
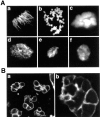
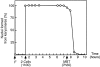
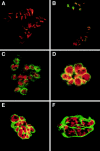
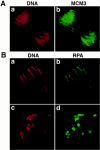
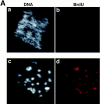
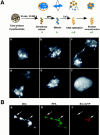
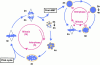
During Xenopus laevis early development, the genome is replicated in less than 15 min every 30 min. We show that during this period, DNA replication proceeds in an atypical manner. Chromosomes become surrounded by a nuclear membrane lamina forming micronuclei or karyomeres. This genomic organization permits that prereplication centers gather on condensed chromosomes during anaphase and that DNA replication initiates autonomously in karyomeres at early telophase before nuclear reconstruction and mitosis completion. The formation of karyomeres is not dependent on DNA replication but requires mitotic spindle formation and the normal segregation of chromosomes. Thus, during early development, chromosomes behave as structurally and functionally independent units. The formation of a nuclear envelope around each chromosome provides an in vivo validation of its role in regulating initiation of DNA replication, enabling the rate of replication to accelerate and S phase to overlap M phase without illegitimate reinitiation. The abrupt disappearance of this atypical organization within one cell cycle after thirteen divisions defines a novel developmental transition at the blastula stage, which may affect both the replication and the transcription programs of development.
Figures


References
-
- Baker TA, Bell SP. Polymerases and the replisome: machines within machines. Cell. 1998;92:295–305. - PubMed
-
- Blow JJ, Laskey RA. Initiation of DNA replication in nuclei and purified DNA by a cell-free extract of Xenopuseggs. Cell. 1986;47:577–587. - PubMed
-
- Blow JJ, Laskey RA. A role for the nuclear envelope in controlling DNA replication within the cell cycle. Nature. 1988;332:546–548. - PubMed
-
- Bravo R, Frank R, Blundell PA, Macdonald-Bravo H. Cyclin/ PCNA is the auxiliary protein of DNA polymerase-delta. Nature. 1987;326:515–517. - PubMed

