Homologous pairing in stretched supercoiled DNA
- PMID: 9724746
- PMCID: PMC27937
- DOI: 10.1073/pnas.95.18.10579
Homologous pairing in stretched supercoiled DNA
Abstract
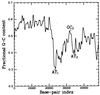
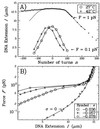
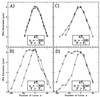
By using elastic measurements on single DNA molecules, we show that stretching a negatively supercoiled DNA activates homologous pairing in physiological conditions. These experiments indicate that a stretched unwound DNA locally denatures to alleviate the force-driven increase in torsional stress. This is detected by hybridization with 1 kb of homologous single-stranded DNA probes. The stretching force involved (approximately 2 pN) is small compared with those typically developed by molecular motors, suggesting that this process may be relevant to DNA processing in vivo. We used this technique to monitor the progressive denaturation of DNA as it is unwound and found that distinct, stable denaturation bubbles formed, beginning in A+T-rich regions.
Figures


Similar articles
-
Supercoiling induces denaturation bubbles in circular DNA.Phys Rev Lett. 2010 Nov 12;105(20):208101. doi: 10.1103/PhysRevLett.105.208101. Epub 2010 Nov 11. Phys Rev Lett. 2010. PMID: 21231267
-
Force-dependent melting of supercoiled DNA at thermophilic temperatures.Biophys Chem. 2014 Mar-Apr;187-188:23-8. doi: 10.1016/j.bpc.2014.01.001. Epub 2014 Jan 17. Biophys Chem. 2014. PMID: 24486433
-
Unpaired structures in SCA10 (ATTCT)n.(AGAAT)n repeats.J Mol Biol. 2003 Feb 28;326(4):1095-111. doi: 10.1016/s0022-2836(03)00037-8. J Mol Biol. 2003. PMID: 12589756
-
Denaturation transition of stretched DNA.Biochem Soc Trans. 2013 Apr;41(2):639-45. doi: 10.1042/BST20120298. Biochem Soc Trans. 2013. PMID: 23514169 Review.
-
The interactions of enzyme and chemical probes with inverted repeats in supercoiled DNA.J Biomol Struct Dyn. 1983 Oct;1(1):169-82. doi: 10.1080/07391102.1983.10507433. J Biomol Struct Dyn. 1983. PMID: 6401110 Review.
Cited by
-
Single-molecule insight into stalled replication fork rescue in Escherichia coli.Nucleic Acids Res. 2021 May 7;49(8):4220-4238. doi: 10.1093/nar/gkab142. Nucleic Acids Res. 2021. PMID: 33744948 Free PMC article. Review.
-
Coexistence of twisted, plectonemic, and melted DNA in small topological domains.Biophys J. 2014 Mar 4;106(5):1174-81. doi: 10.1016/j.bpj.2014.01.017. Biophys J. 2014. PMID: 24606941 Free PMC article.
-
Physiological levels of salt and polyamines favor writhe and limit twist in DNA.Macromolecules. 2012 Apr 10;45(7):3188-3196. doi: 10.1021/ma300211t. Epub 2012 Mar 30. Macromolecules. 2012. PMID: 23526178 Free PMC article.
-
Quantitative modeling and optimization of magnetic tweezers.Biophys J. 2009 Jun 17;96(12):5040-9. doi: 10.1016/j.bpj.2009.03.055. Biophys J. 2009. PMID: 19527664 Free PMC article.
-
Single-molecule force spectroscopy: optical tweezers, magnetic tweezers and atomic force microscopy.Nat Methods. 2008 Jun;5(6):491-505. doi: 10.1038/nmeth.1218. Nat Methods. 2008. PMID: 18511917 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

