Nucleolar localization of early tRNA processing
- PMID: 9716399
- PMCID: PMC317091
- DOI: 10.1101/gad.12.16.2463
Nucleolar localization of early tRNA processing
Abstract
There is little information as to the location of early tRNA biosynthesis. Using fluorescent in situ hybridization in the budding yeast, Saccharomyces cerevisiae, examples of nuclear pre-tRNAs are shown to reside primarily in the nucleoli. We also probed the RNA subunit of RNase P. The majority of the signal from RNase P probes was nucleolar, with less intense signals in the nucleoplasm. These results demonstrate that a major portion of the tRNA processing pathway is compartmentalized in nucleoli with rRNA synthesis and ribosomal assembly. The spatial juxtaposition suggests the possibility of direct coordination between tRNA and ribosome biosynthesis.
Figures

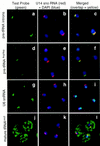
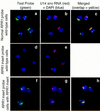
Similar articles
-
Rpp1, an essential protein subunit of nuclear RNase P required for processing of precursor tRNA and 35S precursor rRNA in Saccharomyces cerevisiae.Genes Dev. 1997 Nov 1;11(21):2926-37. doi: 10.1101/gad.11.21.2926. Genes Dev. 1997. PMID: 9353260 Free PMC article.
-
RNase MRP and rRNA processing.Mol Biol Rep. 1995-1996;22(2-3):69-73. doi: 10.1007/BF00988708. Mol Biol Rep. 1995. PMID: 8901490 Review.
-
An RNase P RNA subunit mutation affects ribosomal RNA processing.Nucleic Acids Res. 1996 Aug 15;24(16):3158-66. doi: 10.1093/nar/24.16.3158. Nucleic Acids Res. 1996. PMID: 8774895 Free PMC article.
-
An essential protein-binding domain of nuclear RNase P RNA.RNA. 2001 Apr;7(4):565-75. doi: 10.1017/s1355838201001996. RNA. 2001. PMID: 11345435 Free PMC article.
-
tRNA transfers to the limelight.Genes Dev. 2003 Jan 15;17(2):162-80. doi: 10.1101/gad.1049103. Genes Dev. 2003. PMID: 12533506 Review. No abstract available.
Cited by
-
TFIIIC localizes budding yeast ETC sites to the nuclear periphery.Mol Biol Cell. 2012 Jul;23(14):2741-54. doi: 10.1091/mbc.E11-04-0365. Epub 2012 Apr 11. Mol Biol Cell. 2012. PMID: 22496415 Free PMC article.
-
The Saccharomyces cerevisiae TRT2 tRNAThr gene upstream of STE6 is a barrier to repression in MATalpha cells and exerts a potential tRNA position effect in MATa cells.Nucleic Acids Res. 2004 Sep 30;32(17):5206-13. doi: 10.1093/nar/gkh858. Print 2004. Nucleic Acids Res. 2004. PMID: 15459290 Free PMC article.
-
Conventional and nonconventional roles of the nucleolus.Int Rev Cytol. 2002;219:199-266. doi: 10.1016/s0074-7696(02)19014-0. Int Rev Cytol. 2002. PMID: 12211630 Free PMC article. Review.
-
Stochastic transcription in the p53-mediated response to DNA damage is modulated by burst frequency.Mol Syst Biol. 2019 Dec;15(12):e9068. doi: 10.15252/msb.20199068. Mol Syst Biol. 2019. PMID: 31885199 Free PMC article.
-
Defects in tRNA processing and nuclear export induce GCN4 translation independently of phosphorylation of the alpha subunit of eukaryotic translation initiation factor 2.Mol Cell Biol. 2000 Apr;20(7):2505-16. doi: 10.1128/MCB.20.7.2505-2516.2000. Mol Cell Biol. 2000. PMID: 10713174 Free PMC article.
References
-
- Carter KC, Bowman D, Carrington W, Fogarty K, McNeil JA, Fay FS, Lawrence JB. A three-dimensional view of precursor messenger RNA metabolism within the mammalian nucleus. Science. 1993;259:1330–1335. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
