Sir proteins, Rif proteins, and Cdc13p bind Saccharomyces telomeres in vivo
- PMID: 9710643
- PMCID: PMC109144
- DOI: 10.1128/MCB.18.9.5600
Sir proteins, Rif proteins, and Cdc13p bind Saccharomyces telomeres in vivo
Abstract
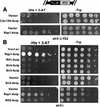
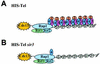
Although a surprisingly large number of genes affect yeast telomeres, in most cases it is not known if their products act directly or indirectly. We describe a one-hybrid assay for telomere binding proteins and use it to establish that six proteins that affect telomere structure or function but which had not been shown previously to bind telomeres in vivo are indeed telomere binding proteins. A promoter-defective allele of HIS3 was placed adjacent to a chromosomal telomere. Candidate proteins fused to a transcriptional activation domain were tested for the ability to activate transcription of the telomere-linked HIS3 gene. Using this system, Rif1p, Rif2p, Sir2p, Sir3p, Sir4p, and Cdc13p were found to be in vivo telomere binding proteins. None of the proteins activated the same reporter gene when it was at an internal site on the chromosome. Moreover, Cdc13p did not activate the reporter gene when it was adjacent to an internal tract of telomeric sequence, indicating that Cdc13p binding was telomere limited in vivo. The amino-terminal 20% of Cdc13p was sufficient to target Cdc13p to a telomere, suggesting that its DNA binding domain was within this portion of the protein. Rap1p, Rif1p, Rif2p, Sir4p, and Cdc13p activated the telomeric reporter gene in a strain lacking Sir3p, which is essential for telomere position effect (TPE). Thus, the telomeric association of these proteins did not require any of the chromatin features necessary for TPE. The data support models in which the telomere acts as an initiation site for TPE by recruiting silencing proteins to the chromosome end.
Figures





References
-
- Alexandre C, Grueneberg D A, Gilman M Z. Studying heterologous transcription factors in yeast. Methods Companion Methods Enzymol. 1993;5:147–155.
-
- Aparicio O M, Billington B L, Gottschling D E. Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae. Cell. 1991;66:1279–1287. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
