Sendai virus Y proteins are initiated by a ribosomal shunt
- PMID: 9710586
- PMCID: PMC109087
- DOI: 10.1128/MCB.18.9.5021
Sendai virus Y proteins are initiated by a ribosomal shunt
Abstract
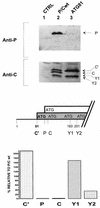
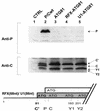
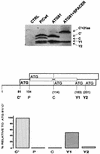
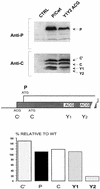
The Sendai virus P/C mRNA expresses eight primary translation products by using a combination of ribosomal choice and cotranscriptional mRNA editing. The longest open reading frame (ORF) of the mRNA starts at AUG104 (the second initiation site) and encodes the 568-amino-acid P protein, an essential subunit of the viral polymerase. The first (ACG81), third (ATG114), fourth (ATG183), and fifth (ATG201) initiation sites are used to express a C-terminal nested set of polypeptides (collectively named the C proteins) in the +1 ORF relative to P, namely, C', C, Y1, and Y2, respectively. Leaky scanning accounts for translational initiation at the first three start sites (a non-ATG followed by ATGs in progressively stronger contexts). Consistent with this, changing ACG81/C' to ATG (GCCATG81G) abrogates expression from the downstream ATG104/P and ATG114/C initiation codons. However, expression of the Y1 and Y2 proteins remains normal in this background. We now have evidence that initiation from ATG183/Y1 and ATG201/Y2 takes place via a ribosomal shunt or discontinuous scanning. Scanning complexes appear to assemble at the 5' cap and then scan ca. 50 nucleotides (nt) of the 5' untranslated region before being translocated to an acceptor site at or close to the Y initiation codons. No specific donor site sequences are required, and translation of the Y proteins continues even when their start codons are changed to ACG. Curiously, ATG codons (in good contexts) in the P ORF, placed either 16 nt upstream of Y1, 29 nt downstream of Y2, or between the Y1 and Y2 codons, are not expressed even in the ACGY1/ACGY2 background. This indicates that ATG183/Y1 and ATG201/Y2 are privileged start sites within the acceptor site. Our observations suggest that the shunt delivers the scanning complex directly to the Y start codons.
Figures



Similar articles
-
Scanning independent ribosomal initiation of the Sendai virus Y proteins in vitro and in vivo.EMBO J. 1989 Feb;8(2):521-6. doi: 10.1002/j.1460-2075.1989.tb03406.x. EMBO J. 1989. PMID: 2542021 Free PMC article.
-
Identification of a cis-acting element required for shunt-mediated translational initiation of the Sendai virus Y proteins.Nucleic Acids Res. 2003 Jan 15;31(2):608-18. doi: 10.1093/nar/gkg143. Nucleic Acids Res. 2003. PMID: 12527769 Free PMC article.
-
Ribosomal initiation from an ACG codon in the Sendai virus P/C mRNA.EMBO J. 1988 Jan;7(1):245-51. doi: 10.1002/j.1460-2075.1988.tb02806.x. EMBO J. 1988. PMID: 2834203 Free PMC article.
-
Sendai virus C proteins are categorically nonessential gene products but silencing their expression severely impairs viral replication and pathogenesis.Genes Cells. 1998 Feb;3(2):111-24. doi: 10.1046/j.1365-2443.1998.00170.x. Genes Cells. 1998. PMID: 9605405
-
Structural and mechanistic insights into hepatitis C viral translation initiation.Nat Rev Microbiol. 2007 Jan;5(1):29-38. doi: 10.1038/nrmicro1558. Epub 2006 Nov 27. Nat Rev Microbiol. 2007. PMID: 17128284 Review.
Cited by
-
A Novel Squirrel Respirovirus with Putative Zoonotic Potential.Viruses. 2018 Jul 18;10(7):373. doi: 10.3390/v10070373. Viruses. 2018. PMID: 30021939 Free PMC article.
-
The amino-terminal extensions of the longer Sendai virus C proteins modulate pY701-Stat1 and bulk Stat1 levels independently of interferon signaling.J Virol. 2003 Feb;77(4):2321-9. doi: 10.1128/jvi.77.4.2321-2329.2003. J Virol. 2003. PMID: 12551969 Free PMC article.
-
Deletion of the D domain of the human parainfluenza virus type 3 (HPIV3) PD protein results in decreased viral RNA synthesis and beta interferon (IFN-β) expression.Virus Genes. 2013 Aug;47(1):10-9. doi: 10.1007/s11262-013-0919-x. Epub 2013 May 18. Virus Genes. 2013. PMID: 23686695
-
Tethering of eIF4G to adenoviral mRNAs by viral 100k protein drives ribosome shunting.Genes Dev. 2004 Aug 15;18(16):1997-2009. doi: 10.1101/gad.1212504. Genes Dev. 2004. PMID: 15314025 Free PMC article.
-
Dengue virus utilizes a novel strategy for translation initiation when cap-dependent translation is inhibited.J Virol. 2006 Mar;80(6):2976-86. doi: 10.1128/JVI.80.6.2976-2986.2006. J Virol. 2006. PMID: 16501107 Free PMC article.
References
-
- Bonneville J-M, Hohn T, Pfeiffer P. Reverse transcription in the plant virus, cauliflower mosaic virus. In: Domingo E, Holland J J, Ahlquist P, editors. RNA Genetics. Boca Raton, Fla: CRC Press; 1988. pp. 23–42.
-
- Chevrier D, Vézina C, Bastille J, Linhard C, Sonenberg N, Boileau G. Higher order structures of the 5′-proximal region decrease the efficiency of the porcine pro-opiomelanocortin mRNA. J Biol Chem. 1988;263:902–910. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
