Sequence analysis of Mus dunni endogenous virus reveals a hybrid VL30/gibbon ape leukemia virus-like structure and a distinct envelope
- PMID: 9696842
- PMCID: PMC109979
- DOI: 10.1128/JVI.72.9.7459-7466.1998
Sequence analysis of Mus dunni endogenous virus reveals a hybrid VL30/gibbon ape leukemia virus-like structure and a distinct envelope
Abstract
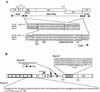
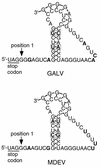
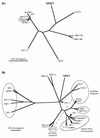
Mus dunni endogenous virus (MDEV) can be activated from M. dunni cells by exposing the cells to hydrocortisone or 5-iodo-2'-deoxyuridine. Interference analysis has revealed that MDEV uses a receptor for cell entry that is different from those used by other murine retroviruses. The entire genome has now been sequenced, revealing a long terminal repeat (LTR)-gag-pol-env-LTR structure typical of simple retroviruses of the murine leukemia virus genus, with no additional open reading frames between env and the 3' LTR. The LTRs and other noncoding regions of MDEV are most closely related to those of VL30 elements, while the majority of the coding sequences are most closely related to those of gibbon ape leukemia virus. MDEV represents the first example of a naturally occurring, replication-competent virus with sequences closely related to VL30 elements. The U3 region of MDEV contains six nearly perfect 80-bp repeats and the beginning of a seventh, and the region expected to contain the packaging sequence contains approximately four imperfect 33-bp repeats. The receptor specificity domains of the envelope are unique among retroviruses and show no apparent similarity to regions of known proteins.
Figures

Similar articles
-
Replication of Mus dunni endogenous retrovirus depends on promoter activation followed by enhancer multimerization.J Virol. 1999 Dec;73(12):9803-9. doi: 10.1128/JVI.73.12.9803-9809.1999. J Virol. 1999. PMID: 10559291 Free PMC article.
-
Molecular cloning of Mus dunni endogenous virus: an unusual retrovirus in a new murine viral interference group with a wide host range.J Virol. 1997 Jun;71(6):4663-70. doi: 10.1128/JVI.71.6.4663-4670.1997. J Virol. 1997. PMID: 9151860 Free PMC article.
-
Genetic organization of gibbon ape leukemia virus.Virology. 1989 Nov;173(1):205-13. doi: 10.1016/0042-6822(89)90236-5. Virology. 1989. PMID: 2683360
-
Molecular biology of type A endogenous retrovirus.Kitasato Arch Exp Med. 1990 Sep;63(2-3):77-90. Kitasato Arch Exp Med. 1990. PMID: 1710682 Review.
-
HIV-1 replication.Somat Cell Mol Genet. 2001 Nov;26(1-6):13-33. doi: 10.1023/a:1021070512287. Somat Cell Mol Genet. 2001. PMID: 12465460 Review.
Cited by
-
Identification of a group of Mus dunni endogenous virus-like endogenous retroviruses from the C57BL/6J mouse genome: proviral genomes, strain distribution, expression characteristics, and genomic integration profile.Chromosome Res. 2012 Oct;20(7):859-74. doi: 10.1007/s10577-012-9322-z. Epub 2012 Nov 30. Chromosome Res. 2012. PMID: 23197326 Free PMC article.
-
Identification of an internal ribosome entry segment in the 5' region of the mouse VL30 retrotransposon and its use in the development of retroviral vectors.J Virol. 1999 Oct;73(10):8393-402. doi: 10.1128/JVI.73.10.8393-8402.1999. J Virol. 1999. PMID: 10482590 Free PMC article.
-
Replication of Mus dunni endogenous retrovirus depends on promoter activation followed by enhancer multimerization.J Virol. 1999 Dec;73(12):9803-9. doi: 10.1128/JVI.73.12.9803-9809.1999. J Virol. 1999. PMID: 10559291 Free PMC article.
-
Characterization of endogenous retroviruses in sheep.J Virol. 2003 Oct;77(20):11268-73. doi: 10.1128/jvi.77.20.11268-11273.2003. J Virol. 2003. PMID: 14512574 Free PMC article.
-
Novel endogenous type C retrovirus in baboons: complete sequence, providing evidence for baboon endogenous virus gag-pol ancestry.J Virol. 1999 Aug;73(8):7021-6. doi: 10.1128/JVI.73.8.7021-7026.1999. J Virol. 1999. PMID: 10400802 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

